Teresita Porter 🙋🏻♀️ · @DNAdataPhile
145 followers · 531 posts · Server ecoevo.socialIntragenomic diversity of the V9 hypervariable domain in eukaryotes has little effect on metabarcoding
https://www.cell.com/iscience/pdf/S2589-0042(23)01368-8.pdf
#18S #v9 #IntragenomicVariation #DADA2 #SWARM #metabarcoding
#18S #v9 #intragenomicvariation #dada2 #swarm #Metabarcoding
Teresita Porter 🙋🏻♀️ · @DNAdataPhile
116 followers · 433 posts · Server ecoevo.social"Evaluating the diversity of the enigmatic fungal phylum Cryptomycota across habitats using 18S rRNA metabarcoding"
https://www.sciencedirect.com/science/article/abs/pii/S1754504823000259
#cryptomycota #18S #metabarcoding #diversity #fungi #mycology
#cryptomycota #18S #Metabarcoding #diversity #fungi #mycology
IMPACTT Microbiome · @IMPACTT
75 followers · 30 posts · Server mstdn.scienceToday is the LAST DAY to apply for our joint CBW-IMPACTT Microbiome Analysis Workshop at the early-bird rate! July 5-7, 2023 at the University of Calgary. Apply here: https://bioinformatics.ca/workshops-all/2023-cbw-impactt-microbiome-analysis/
#bioinformatics #workshop #16S #18S #sequencing #shotgunsequencing #metagenomics #genomics #transcriptomics #metatranscriptomics #longread #markergene #functionalannotation #tutorial #statistics #biostatistics #microbiome #microbiota #microbialecology #diversity #betadiversity #differentialabundance
#bioinformatics #workshop #16s #18S #sequencing #shotgunsequencing #Metagenomics #genomics #Transcriptomics #metatranscriptomics #longread #markergene #functionalannotation #Tutorial #statistics #biostatistics #microbiome #microbiota #microbialecology #diversity #betadiversity #differentialabundance
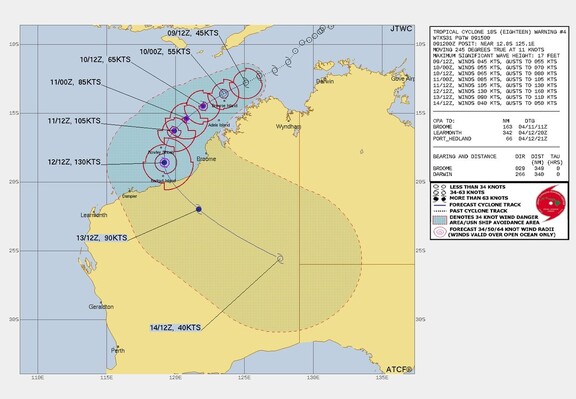
Zoom Earth · @zoom_earth
723 followers · 177 posts · Server mapstodon.spaceSunrise over Tropical Cyclone #18S near Western Australia. Winds reaching 95 km/h.
Zoom Earth · @zoom_earth
721 followers · 176 posts · Server mapstodon.spaceZoom Earth · @zoom_earth
721 followers · 175 posts · Server mapstodon.spaceSunrise over Tropical Cyclone #18S near Western Australia. Winds close to 95 km/h.
Brandon · @b3108
7 followers · 1566 posts · Server mastodon.worldZoom Earth · @zoom_earth
721 followers · 174 posts · Server mapstodon.spaceZoom Earth · @zoom_earth
719 followers · 173 posts · Server mapstodon.spaceZoom Earth · @zoom_earth
719 followers · 172 posts · Server mapstodon.spaceTropical Cyclone #18S has been designated in the Timor Sea.
Follow updates in real-time here: https://zoom.earth/storms/18s-2023/
marcgarel (PhD) · @marcgarel
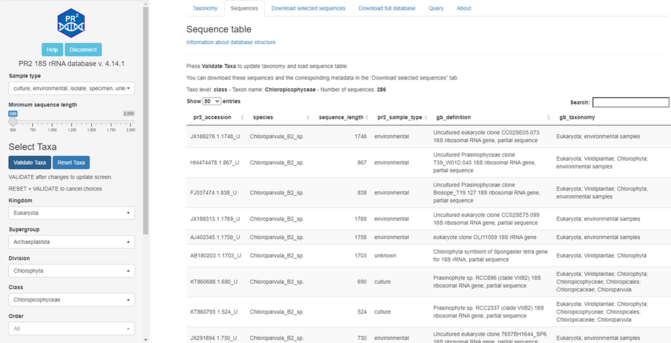
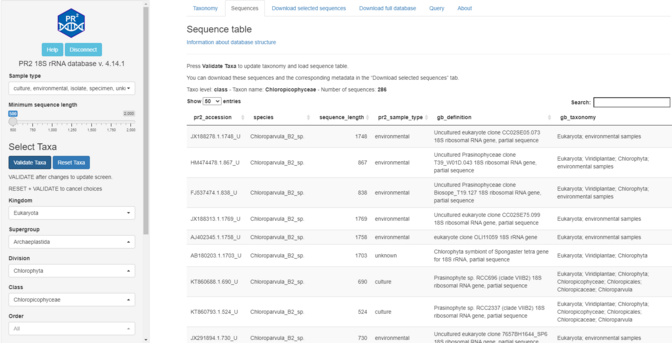
24 followers · 32 posts · Server ecoevo.socialRT @PR2database Announcing the release of version 4.14.1 of the PR2 database of 18S eukaryotic reference sequences.
The database itself is unchanged but we now provide a web interface:
app.pr2-database.org
More information: https://pr2database.github.io/pr2database/articles/vignette-shiny-presentation.html. #protists #metabarcoding #database #18S.
#protists #Metabarcoding #database #18S
DudinLab · @DudinLab
643 followers · 119 posts · Server mstdn.socialRT @PR2database
Announcing the release of version 4.14.1 of the PR2 database of 18S eukaryotic reference sequences.
The database itself is unchanged but we now provide a web interface:
More information: https://pr2database.github.io/pr2database/articles/vignette-shiny-presentation.html
#18S #database #metabarcoding #protists
DEEM team Orsay · @DEEMteam
30 followers · 7 posts · Server ecoevo.socialRT @PR2database@twitter.com
Announcing the release of version 4.14.1 of the PR2 database of 18S eukaryotic reference sequences.
The database itself is unchanged but we now provide a web interface:
More information: https://pr2database.github.io/pr2database/articles/vignette-shiny-presentation.html
#protists #metabarcoding #database #18S
🐦🔗: https://twitter.com/PR2database/status/1596067802216992768
#protists #Metabarcoding #database #18S
Daniel Vaulot 🇪🇺🇺🇦 · @daniel_vaulot
129 followers · 170 posts · Server genomic.socialAnnouncing the release of version 4.14.1 of the PR2 database of 18S eukaryotic reference sequences.
The database itself is unchanged, but we now provide a web interface:
More information: https://pr2database.github.io/pr2database/articles/vignette-shiny-presentation.html
#protists #Metabarcoding #database #18S
Lucie Bittner · @luciebittner
58 followers · 32 posts · Server ecoevo.socialRT @PR2database
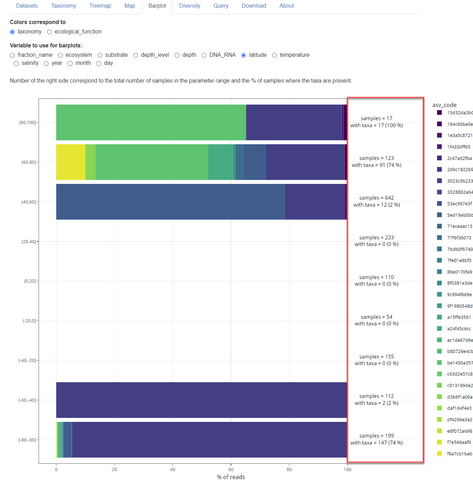
Announcing the release of version 2.0 of metaPR2 database of 18S eukaryotic metabarcodes
#protists #Metabarcoding #biogeography #18S
Daniel Vaulot 🇪🇺🇺🇦 · @daniel_vaulot
123 followers · 154 posts · Server genomic.socialAnnouncing the release of version 2.0 of metaPR2, database of 18S eukaryotic metabarcodes
https://shiny.metapr2.org/metapr2/
Major changes are listed here: https://pr2database.github.io/metapr2-shiny/news/index.html
* Addition of 18 new datasets
* Use of clustered metabarcodes (ASVs)
* New panel listing taxonomy of metabarcodes
* Barplots now include samples where taxa are absent
Please report any issue here: https://matrix.to/#/#pr2-database:matrix.org
#protists #Metabarcoding #biogeography #18S
Daniel Vaulot 🇪🇺🇺🇦 · @daniel_vaulot
123 followers · 153 posts · Server genomic.socialNew paper on #foraminfera making use of the #pr2database.
#foraminfera #pr2database #protists #Metabarcoding #18S
Daniel Vaulot · @daniel_vaulot
76 followers · 75 posts · Server genomic.socialIf you are interested in hearing the latest news and development of the PR2 database ecosystem for 18S rRNA metabarcoding and/or contribute to PR2, please join us at
Daniel Vaulot · @daniel_vaulot
76 followers · 75 posts · Server genomic.socialEukRibo a new database of 18S sequences developed from the PR2 database
laseab · @laseab
32 followers · 6 posts · Server genomic.socialUsually working with sequencing 16S but I want to try out 18S with a student coming in. We would like to try to find primer/primer combinations which work best for different environments we are working in, e.g. sediment, biofilm, baltic sea water and deep biosphere (low yield of DNA). We have a bunch of primers and set ups at our department, which we will try out, but I thought I would give it a shot here as well. So if anyone has any recommendations:) Thanks #18S #sequencing #TipsAndTricks
#18S #sequencing #TipsAndTricks