LBNL BioSci · @LBNLBioSci
103 followers · 160 posts · Server sciencemastodon.comUsing X-ray crystallography at the Advanced Light Source (ALS), research scientist Banumathi Sankaran at the Berkeley Center for Structural Biology (BCSB) has helped confirm the successful design of proteins that toggle between two different shapes in response to biological triggers.
Berkeley Lab #ProteinDesign #MachineLearning #StructuralBiology #Science #RoseTTAFold #ProteinMPNN #AlphaFold2
#proteindesign #MachineLearning #structuralbiology #science #RoseTTAFold #proteinmpnn #AlphaFold2
CellBioNews · @cellbionews
57 followers · 1363 posts · Server scientificnetwork.de#Symbiotic and #pathogenic #fungi may use similar molecular tools to manipulate #plants.
#mycorrhiza #AlphaFold2 #AI #Rhizophagus #MycFOLDs
https://phys.org/news/2023-06-symbiotic-pathogenic-fungi-similar-molecular.html
#symbiotic #pathogenic #fungi #plants #mycorrhiza #AlphaFold2 #ai #rhizophagus #mycfolds
Vaclav Hanzl · @vaclavh
22 followers · 48 posts · Server sigmoid.socialReading #AlphaFold2 and #OMSF #OpenFold code is great but with #protein #molecules in your hands you feel it better. Designing the kit in #FreeCAD ensures you know bond lengths and angles etc. So I spent few days learning and printing. The kit is optimized for protein #folding, single bond rotates but can be fixed in #peptide bond, it keeps shape even before adding #hydrogen #bonds in #alphahelix (or pair #RNA bases by H-bond).
Print your #kit and enjoy #biochemistry!
https://www.printables.com/model/457984-molecules-for-protein-folding
#AlphaFold2 #omsf #openfold #protein #molecules #freecad #folding #peptide #hydrogen #bonds #alphahelix #RNA #kit #biochemistry
Laborjournal · @laborjournal
521 followers · 241 posts · Server mstdn.scienceIm Kielwasser von #AlphaFold2 jagt gegenwärtig ein Programm zur Vorhersage von #Proteinstrukturen das nächste. Was müssen sie mitbringen, um auch die Mechanismen der Proteinfaltung zu enträtseln? …
— Henrik Müller gibt Antworten: 👉 https://www.laborjournal.de/rubric/methoden/methoden/v276.php
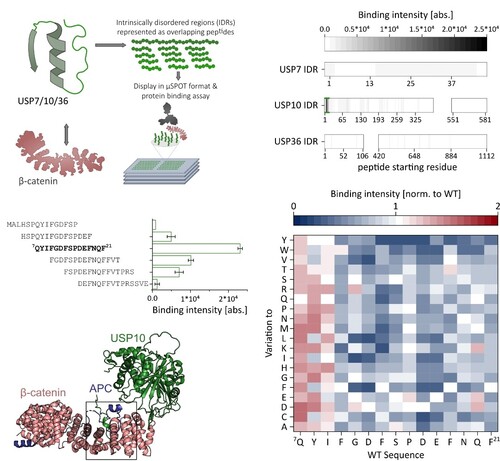
Clemens Schulte · @SchulteClemens
32 followers · 19 posts · Server mstdn.scienceGreat to see this collaborative effort spearheaded by Michaela Reissland from http://diefenbacher-lab.com/ out as #preprint 📝
Stabilisation of β-Catenin-WNT signalling by USP10 in APC-truncated colorectal cancer drives cancer stemness and enables super-competitor signalling
https://www.biorxiv.org/content/10.1101/2023.02.10.527983v1
#peptide microarray studies in our lab and #AlphaFold2 Multimer provided insights into β-catenin - USP10 binding in single amino acid resolution. Huge thx to all co-authors! 👏 #cancer #research
#preprint #peptide #AlphaFold2 #cancer #research
PLOS Biology · @PLOSBiology
4717 followers · 833 posts · Server fediscience.orgHallvard Olsvik & @TerjeJohansen17 explore a new #PLOSBiology study showing how the AI-based tool #AlphaFold2-multimer can be used to identify sequence motifs that bind to the ATG8 family of #autophagy proteins. Paper: https://plos.io/3YDtLIM Primer: https://plos.io/40TgKwG
#autophagy #AlphaFold2 #plosbiology
PLOS Biology · @PLOSBiology
4715 followers · 825 posts · Server fediscience.org#Alphafold2-multimer assisted prediction of typical & atypical ATG8-binding motifs enables fast-forward discovery of #autophagy modulators, including the central autophagy regulator ATG3 @TarhanEI @BubeckLab @Tolga_Bzkrt &co #PLOSBiology https://plos.io/3YDtLIM
#plosbiology #autophagy #AlphaFold2
PLOS Biology · @PLOSBiology
4715 followers · 825 posts · Server fediscience.orgHallvard Olsvik & @TerjeJohansen17 explore a new #PLOSBiology study showing how the AI-based tool #AlphaFold2-multimer can be used to identify sequence motifs that bind to the ATG8 family of #autophagy proteins. Paper: https://plos.io/3YDtLIM Primer: https://plos.io/40TgKwG
#autophagy #AlphaFold2 #plosbiology
PLOS Biology · @PLOSBiology
4715 followers · 825 posts · Server fediscience.org#Alphafold2-multimer assisted prediction of typical & atypical ATG8-binding motifs enables fast-forward discovery of #autophagy modulators, including the central autophagy regulator ATG3 @TarhanEI @BubeckLab @Tolga_Bzkrt &co #PLOSBiology https://plos.io/3YDtLIM
#plosbiology #autophagy #AlphaFold2
PLOS Biology · @PLOSBiology
4715 followers · 819 posts · Server fediscience.orgHallvard Olsvik & @TerjeJohansen17 explore a new #PLOSBiology study showing how the AI-based tool #AlphaFold2-multimer can be used to identify sequence motifs that bind to the ATG8 family of #autophagy proteins. Paper: https://plos.io/3YDtLIM Primer: https://plos.io/40TgKwG
#autophagy #AlphaFold2 #plosbiology
PLOS Biology · @PLOSBiology
4715 followers · 820 posts · Server fediscience.orgRT @TarhanEI
Officially out in @PLOSBiology!! Take a look at how we are accelerating our research with AI @DeepMind #AlphaFold2 https://twitter.com/PLOSBiology/status/1623622070226632705
Jamie H D Cate · @jhdcate
340 followers · 514 posts · Server fediscience.org@holeungng I think this paper makes a strong case for using perturbation analysis like adversarial attacks to test for the robustness of protein folding neural networks (PFNNs). They conclude AF2 is not robust enough yet to say that the protein fold prediction problem has been solved. #AlphaFold2 #AI
Andras Toth · @tothur
148 followers · 209 posts · Server mastodon.socialA fehérjék szekvenciájából 3D szerkezeteket előállító #AlphaFold2 2020-ban megoldotta a molekuláris biológia egyik legnagyobb problémáját.
Az algoritmusok legújabb versenyén az is körvonalazódott, hogy merre halad tovább a terület. #AI
- erről is írtam a héten:
Alexis Verger · @AVerger
1147 followers · 154 posts · Server fediscience.org👀 #Preprint @strucbio
#AlphaFold2 can predict #structural and #phenotypic effects of #single #mutations https://www.biorxiv.org/content/10.1101/2022.04.14.488301v2
#mutations #single #phenotypic #structural #AlphaFold2 #preprint
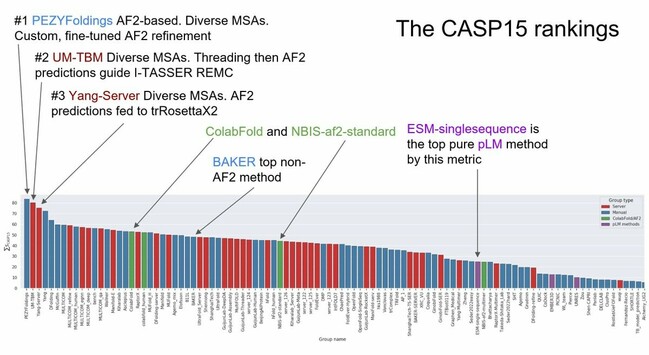
Albert Vilella · @albertvilella
345 followers · 199 posts · Server genomic.socialMore notes from the #CASP15 competition. Other resources here: https://www.aditiashenoy.com/posts/casp15/day2/
RT @AlbertVilella@twitter.com
Some initial results from the #CASP15 competition and we see that #Alphafold2 has become a fertile ground for experimentation by several research groups around the world. Facebook's ESM protein language models (pLMs) are the top non-MSA based methods.
🐦🔗: https://twitter.com/AlbertVilella/status/1601909663309901824
Juan Mateos-Garcia · @jmateosg
235 followers · 134 posts · Server econtwitter.netStanding on the shoulders of giant neural networks :)
RT @AlbertVilella@twitter.com
Some initial results from the #CASP15 competition and we see that #Alphafold2 has become a fertile ground for experimentation by several research groups around the world. Facebook's ESM protein language models (pLMs) are the top non-MSA based methods.
🐦🔗: https://twitter.com/AlbertVilella/status/1601909663309901824
Gemma C. Atkinson · @Gemma_Atkinson
268 followers · 63 posts · Server fediscience.orglast week to apply!
RT @gem__atkinson@twitter.com
Do you love structural #bioinformatics? Would you like to be based in a beautiful city & region, working with colleagues to make exciting new discoveries? Consider joining us in Lund University https://lu.varbi.com/en/what:job/jobID:564808/
#structuralbiology #alphafold2 #alphafold #AI
Please share!
🐦🔗: https://twitter.com/gem__atkinson/status/1598234355859881984
#ai #alphafold #AlphaFold2 #StructuralBiology #bioinformatics
Gemma C. Atkinson · @Gemma_Atkinson
154 followers · 40 posts · Server fediscience.org***One week left to apply!***
Do you love structural #bioinformatics? Would you like to be based in a beautiful city & region, working with colleagues to make exciting new discoveries? Consider joining us in Lund University https://lu.varbi.com/en/what:job/jobID:564808/
#structuralbiology #alphafold2 #alphafold #AI #cryoEM #computationalbiology
Please share!
#computationalbiology #CryoEM #ai #alphafold #AlphaFold2 #StructuralBiology #bioinformatics
Dave Briggs :protein: · @xtaldave
1019 followers · 2278 posts · Server xtaldave.netThis looks useful for generating alternative conformations of various classes of proteins for drug design.
Biasing AlphaFold2 to predict GPCRs and Kinases with user-defined functional or structural properties
#DrugDesign #StrucuralBiology @strucbio #science #AlphaFold2
http://biorxiv.org/cgi/content/short/2022.12.11.519936v1?rss=1
#DrugDesign #strucuralbiology #Science #AlphaFold2
Juan Mateos-Garcia · @jmateosg
235 followers · 134 posts · Server econtwitter.netRT @AlbertVilella@twitter.com
Some initial results from the #CASP15 competition and we see that #Alphafold2 has become a fertile ground for experimentation by several research groups around the world. Facebook's ESM protein language models (pLMs) are the top non-MSA based methods.
🐦🔗: https://twitter.com/AlbertVilella/status/1601909663309901824