Albert Cardona · @albertcardona
1978 followers · 3594 posts · Server mathstodon.xyzMay I suggest #FijiSc https://fiji.sc which is a free and open source image processing software. Handling very large images should not be an issue. We handle volumes of many terabytes with it. Can open and write pretty much any file format imaginable, including raw, and offers a huge amount of image filters.
Albert Cardona · @albertcardona
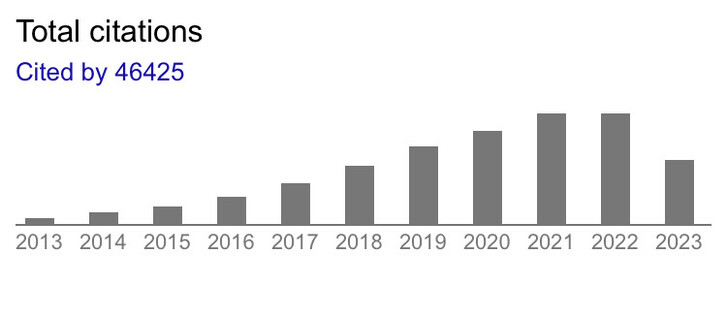
1906 followers · 3298 posts · Server mathstodon.xyz@quantixed Coincidentally, this year the #FijiSc software paper’s citations plateaued, after 10 years of growth (published in 2012 https://www.nature.com/articles/nmeth.2019 ), as if there weren’t any more labs that could start to cite it. Knowing the size of biology academia would be interesting.
Albert Cardona · @albertcardona
1843 followers · 3056 posts · Server mathstodon.xyz@philiphubbard Have you tried Benjamin Schmid's 3D animation and rendering language, #3DScript, for #FijiSc:
https://bene51.github.io/3Dscript/
It's really not hard and surprisingly capable.
There's also a paper about it: https://www.nature.com/articles/s41592-019-0359-1
#3d #imageprocessing #FijiSc #3dscript
Albert Cardona · @albertcardona
1841 followers · 3038 posts · Server mathstodon.xyzDeepImageJ 3.0 released—a user-friendly #FijiSc plugin that enables the use of a variety of pre-trained neural networks from the #BioimageModelZoo. Works with small and also very large images thanks to #ImgLib2.
#imageprocessing #imagej #tensorflow #pytorch #deepimagej #ImgLib2 #bioimagemodelzoo #FijiSc
Albert Cardona · @albertcardona
1837 followers · 3022 posts · Server mathstodon.xyz@ftrain Check out the open source Fiji image processing software https://fiji.sc and the image processing forum https://forum.image.sc
Albert Cardona · @albertcardona
1703 followers · 1966 posts · Server mathstodon.xyzTurns out, this is the result of an artifact: I had transform the images using integer coordinates rather than subpixel floating-point coordinates – my error. Hence these neatly drawn black spaces with sharp boundaries. The scripts have been corrected now; if you want to deliberately recreate this, for artistic purposes, then check the post: https://forum.image.sc/t/any-advice-on-making-a-thin-plate-spline-transform-more-precise/79595/3
#FijiSc
Albert Cardona · @albertcardona
1703 followers · 1963 posts · Server mathstodon.xyzThe underlying source code is here, if you want to try registering pairs of images with a non-linear transformation such as a thin-plate spline, on the bases of blockmatching features:
#java #imageprocessing #jython #FijiSc
Albert Cardona · @albertcardona
1697 followers · 1944 posts · Server mathstodon.xyzEvery time I need to run an image processing task, the open source software #FijiSc delivers.
The javadocs are up to date https://javadoc.scijava.org/ , the libraries just work – particularly #ImgLib2 https://imagej.net/libs/imglib2/
And the examples of my own tutorial https://syn.mrc-lmb.cam.ac.uk/acardona/fiji-tutorial/ written in python 2.7 for the #JVM (#jython), despite some being a decade old, they all just work. Grateful every day for the outstanding backwards compatibility plus the new plugins and libraries that continue to grow https://fiji.sc
Many thanks to the many, many developers and maintainers, particularly Curtis Rueden, who is presently cutting out a new release: 2.11.0 https://forum.image.sc/t/plugin-maintainers-can-you-test-fiji-2-11-0/78852/18
#imageprocessing #jython #jvm #ImgLib2 #FijiSc
Albert Cardona · @albertcardona
1622 followers · 1404 posts · Server mathstodon.xyz@hynek Same here—my best online posts are notes to future self, for e.g., image processing code snippets I don’t want to relearn. Here, some in #jython driving #FijiSc’s #java libraries to register images, transform N-dimensional images, visualise them in 2d/3d/4d, make GUIs, and more: https://syn.mrc-lmb.cam.ac.uk/acardona/fiji-tutorial I hear many found these useful.
Albert Cardona · @albertcardona
1610 followers · 1374 posts · Server mathstodon.xyz@florianjug @humantechnopole @AI4Life
Much envy, wish I was there. Been wanting to bring deep learning to some old corners of #FijiSc
Albert Cardona · @albertcardona
1610 followers · 1374 posts · Server mathstodon.xyz@markkitti Wonderful. Is anyone going to test out deeplearning4j and how to use it from the Weka library, and maybe add a working example for #FijiSc's LabKit? Would be so useful to have this documented.
Albert Cardona · @albertcardona
1367 followers · 723 posts · Server mathstodon.xyz@alexwild @stevenbodzin Agree stacking is great in principle and often poor in practice. In my limited experience, stacking works best with great illumination (full sun, given no light equipment budget) and very short exposure times, and when managed directly by the camera as a brief burst (my Olympus TG-6 handles this somewhat well).
Ages ago I used #ImageJ’s Extended Depth of Field plugin http://bigwww.epfl.ch/demo/edf/index.html
#photography #FijiSc
Albert Cardona · @albertcardona
1103 followers · 185 posts · Server mathstodon.xyzAlbert Cardona · @albertcardona
946 followers · 4 posts · Server mathstodon.xyzHave you discovered the image processing forum yet? https://forum.image.sc
Featuring tutorials, requests for guidance, help, discussion of new features and documentation for open source software #FijiSc #ImageJ #CellProfiler #DeepLabCut #Icy #ilastik #napari #QuPath #scikitimage #StarDist and many more.
#groovy #kotlin #python #java #StarDist #scikitimage #QuPath #napari #ilastik #icy #DeepLabCut #CellProfiler #imagej #FijiSc
Albert Cardona · @albertcardona
913 followers · 2 posts · Server mathstodon.xyz#TrakEM2 runs as a plugin of #FijiSc https://fiji.sc/ and in fact motivated the creation of the #FijiSc software in the first place, to manage its many dependencies and facilitate distribution to the broader #neuroscience community.
#TrakEM2 was founded in 2005, when TB-sized datasets were rare and considered large. Largest dataset I've successfully managed with #TrakEM2 was ~16 TB. For larger volumes see #CATMAID.
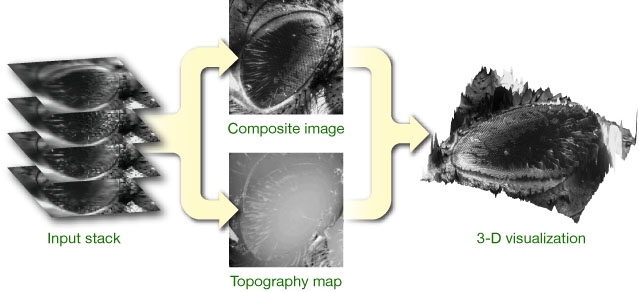
For 3D visualization #TrakEM2 uses the 3D Viewer https://imagej.net/plugins/3d-viewer/
#catmaid #neuroscience #FijiSc #TrakEM2
Laurent Thomas · @LauLauThom
27 followers · 28 posts · Server qoto.org@trinsec yeah let's do some #tootorials ! I might make threads about #FijiSc #bioimageanalysis #microscopy or #diy in the future! Anything like this ThreadReader yet for the fediverse?
#BioImageAnalysis #diy #tootorials #FijiSc #microscopy
Albert Cardona · @albertcardona
697 followers · 557 posts · Server qoto.orgHave you discovered the image processing forum yet? https://forum.image.sc
Featuring tutorials, requests for guidance, help, discussion of new features and documentation for open source software #FijiSc #ImageJ #CellProfiler #DeepLabCut #Icy #ilastik #napari #QuPath #scikitimage #StarDist and many more.
#java #python #kotlin #groovy
#ilastik #StarDist #FijiSc #imagej #napari #QuPath #scikitimage #java #python #kotlin #groovy #CellProfiler #DeepLabCut #icy
Albert Cardona · @albertcardona
697 followers · 557 posts · Server qoto.org@engineeringjoy @Russpoldrack @weinberz @SherlockpHolmes @PracheeAC Welcome fellow #ImageJ -er. Curious about Ca2+ analysis: does it run within ImageJ (or #FijiSc?) and if so which plugin do you use?
Albert Cardona · @albertcardona
697 followers · 557 posts · Server qoto.orgAlbert Cardona · @albertcardona
697 followers · 557 posts · Server qoto.org@somedonkey Beautiful!
By the way this reminded me there's an upcoming paper by J-Y Tinevez and Pavel Tomancak on the open source software Mastodon (they are going to have to find a different hashtag) for #FijiSc that would benefit so much from your beautiful illustrations: https://github.com/mastodon-sc/mastodon
(The Mastodon cell tracking software is an evolution of their former MaMuT software https://imagej.net/plugins/mamut/index )