Albert Cardona · @albertcardona
1846 followers · 3078 posts · Server mathstodon.xyz"A consensus cell type atlas from multiple connectomes reveals principles of circuit stereotypy and variation", by Schlegel et al. 2023 (Greg Jefferis lab at #MRCLMB in collaboration with the Bock, Murthy, and Seung labs, and also Volker Hartenstein)
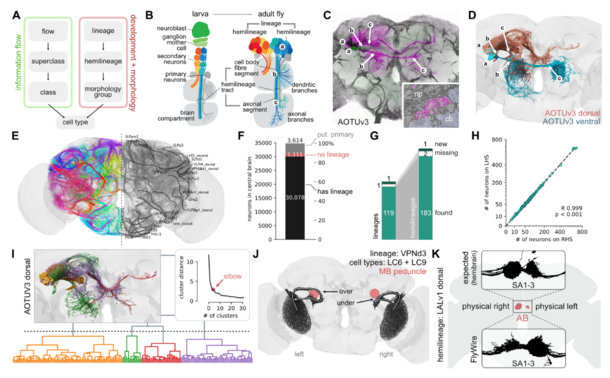
Interesting to me in particular, and here I see the hand of Volker Hartenstein (my postdoc adviser!), is the annotation of developmental units: the neuronal lineages (Fig 2 below).
#devbiol #neuroscience #connectomics #drosophila #MRCLMB
Albert Cardona · @albertcardona
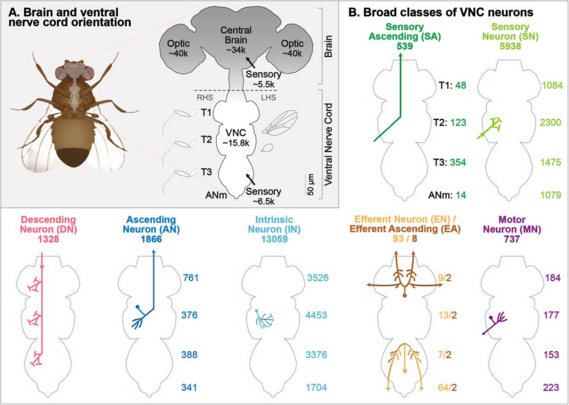
1846 followers · 3077 posts · Server mathstodon.xyz"Systematic annotation of a complete adult male Drosophila nerve cord connectome reveals principles of functional organisation", by Marin et al. 2023 (Greg Jefferis lab at #MRCLMB in collaboration with #HHMIJanelia and others).
https://www.biorxiv.org/content/10.1101/2023.06.05.543407v1
On the analysis of the #Drosophila male adult nerve cord (#MANC), reconstructed from #VolumeEM by Takemura et al. 2023 (preprint still pending to come out).
#connectomics #neuroscience #volumeem #manc #drosophila #HHMIJanelia #MRCLMB
Albert Cardona · @albertcardona
1811 followers · 2850 posts · Server mathstodon.xyz"On being asked what made the LMB such a remarkable place, Max answered: ‘Creativity in science, as in art [referring to the Renaissance in Florence], cannot be organised. It arises spontaneously from individual talent. Well-run laboratories can foster it, but hierarchical organisations, inflexible bureaucratic rules, and mountains of futile paperwork can kill it. Discoveries cannot be planned, they pop up, like Puck, in unexpected corners.’"
From Daniela Rhodes' 2002 piece on #MaxPerutz
https://www.embopress.org/doi/full/10.1093/embo-reports/kvf103
Albert Cardona · @albertcardona
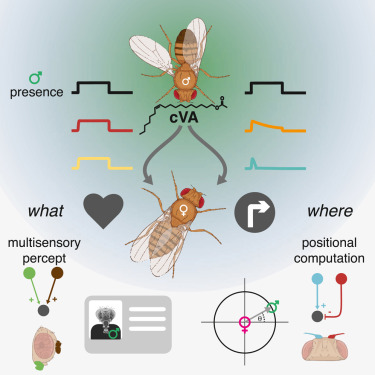
1805 followers · 2680 posts · Server mathstodon.xyz"Generating parallel representations of position and identity in the olfactory system", Taisz et al. 2023 (Greg Jefferis' lab at #MRCLMB) https://www.sciencedirect.com/science/article/pii/S0092867423004725
In a neural circuit of the female brain for processing the scent of the male pheromone (cVA), different features such as that it is a male and where it is in space are read out at different depths of the neural circuit layers.
#connectomics #neuroscience #drosophila #MRCLMB
Albert Cardona · @albertcardona
1689 followers · 1840 posts · Server mathstodon.xyzYou are welcome–it's been fun to discover an entirely different slice of science and research here, starting this past October. My presence in the other side is nominal, keeping an account there as a flag and pointer to here. The move gave me the chance to follow more entomology accounts and learn more about insects and small invertebrates I'd like to map some day. Also: one has to follow more accounts here to sample the #neuroscience and #DevBiol fields as broadly as we did on the other side, as the only promotion algorithm is the boost button.
In other news, this week we installed a Bruker SkyScan microCT in the lab, courtesy of the #MRCLMB (I love this place), and we are over the moon. It's essential to quality-control samples that proceed to electron microscopy for #connectomics. But it's also going to prove useful to scan whole small animals densely at 0.5 to 1 µm isotropic resolution.
#connectomics #MRCLMB #devbiol #neuroscience
Albert Cardona · @albertcardona
1682 followers · 1776 posts · Server mathstodon.xyzIn her book "Ahead of the Curve", Kathleen Weston tells the story of women scientists at the #MRCLMB in Cambridge, UK, such as Barbara Pearse–there's a whole chapter on her fascinating work with clathrin-coated vesicles extracted from pig brains.
Pearse was awarded an EMBO gold medal in 1987 for her pioneering work.
https://www.embopress.org/doi/pdf/10.1002/j.1460-2075.1987.tb02536.x
Albert Cardona · @albertcardona
1639 followers · 1495 posts · Server mathstodon.xyzHarald Hess awarded the 2023 James Prize in Science and Technology Integration by the US National Academy of Sciences: https://www.nasonline.org/programs/awards/2023-awards/Hess.html
At #HHMIJanelia, the lab of Harald Hess revolutionized electron microscopy with enhanced FIBSEM (C. Shan Xu et al. 2017 @eLife https://elifesciences.org/articles/25916 ) –now in use by us at #MRCLMB, and applied for whole #Drosophila CNS imaging; built an automated STEM to image the whole #Drosophila larval body at 5x5x35 nm; and so much more. Hats off! Congratulations!
#drosophila #MRCLMB #HHMIJanelia
Albert Cardona · @albertcardona
1565 followers · 1242 posts · Server mathstodon.xyzFantastic talk today by @DenisDuboule at the #MRCLMB on #Hox genes, highlighting their unique features and introducing the resolution of a long-standing mystery: the Hox timer. As explained in:
"Sequential And Directional Insulation By Conserved CTCF Sites Underlies The Hox Timer In Pseudo-Embryos", Rekaik et al. 2022 https://www.biorxiv.org/content/10.1101/2022.08.29.505673v1
#gastruloids #devbio #hox #MRCLMB
Albert Cardona · @albertcardona
1546 followers · 1161 posts · Server mathstodon.xyz“We had central heating and hot water. They didn’t always work that well but it was way more than what most people had — Americans were told that because of the advantageous English climate you didn’t need heating or refrigeration!”—Joan Steitz, RNA pioneer, discovered the spliceosome https://en.m.wikipedia.org/wiki/Joan_A._Steitz and here commenting on UK housing in the late 1960’s.
From “Ahead of the Curve” by Kathleen Weston, telling the history of the #MRCLMB
#science #academia #history #UK
#uk #history #academia #science #MRCLMB
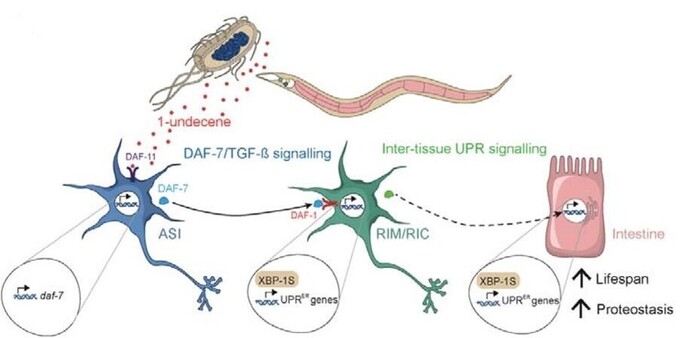
preLights · @preLights
280 followers · 14 posts · Server mstdn.science⚠️ Danger ahead – trust your gut feeling!
#Metabolites from pathogenic #bacteria trigger a #stress response in the worm’s #gut
Great #preLight from #CheeKiangEwe who covers a #preprint from the Taylor lab #MRCLMB. Includes an author's response 📝
#metabolites #bacteria #stress #gut #prelight #cheekiangewe #preprint #MRCLMB
Albert Cardona · @albertcardona
1025 followers · 109 posts · Server mathstodon.xyzA huge THANK YOU to everyone that worked on this project for 10 years, starting with first co-authors Michael Winding and Ben Pedigo at University of Cambridge and Johns Hopkins. A collaboration with Marta Zlatic, Carey E. Priebe, and Joshua Vogelstein.
This work started at #HHMIJanelia and continued at the #MRCLMB in Cambridge, UK.
All neuron reconstructions were done painstakingly by hand with #CATMAID by over >80 people! Thanks so much!
#connectomics #neuroscience #catmaid #MRCLMB #HHMIJanelia
Albert Cardona · @albertcardona
970 followers · 953 posts · Server qoto.orgThe #MRCLMB offers fully funded PhD studentships. Come join us!
Deadline: December 1st, 2022 (six days left).
Have a look at the projects listed for the Neurobiology Division (where Marta Zlatic, Greg Jefferis, Bill Schafer, Marco Tripodi, and many others, including myself).
List of projects: https://www2.mrc-lmb.cam.ac.uk/students/international-phd-programme/projects/
How to apply: https://www2.mrc-lmb.cam.ac.uk/students/international-phd-programme/how-to-apply/
#neuroscience #phd #MRCLMB #academia
Albert Cardona · @albertcardona
697 followers · 557 posts · Server qoto.orgWhat can you do with a #CATMAID server? Say, let's look at the #Drosophila (vinegar fly, often referred to as fruit fly) larval central nervous system, generously hosted by the #VirtualFlyBrain https://l1em.catmaid.virtualflybrain.org/?pid=1&zp=108250&yp=82961.59999999999&xp=54210.799999999996&tool=tracingtool&sid0=1&s0=2.4999999999999996&help=true&layout=h(XY,%20%7B%20type:%20%22neuron-search%22,%20id:%20%22neuron-search-1%22,%20options:%20%7B%22annotation-name%22:%20%22papers%22%7D%7D,%200.6) or the #Platynereis (a marine annelid) server from the Jekely lab https://catmaid.jekelylab.ex.ac.uk/
First, directly interact by point-and-click: open widgets, find neurons by name or annotations, fire up a graph widget and rearrange neurons to make a neat synaptic connectivity diagram, or an adjacency matrix, or look at neuron anatomy in 3D. Most text–names, numbers–are clickable and filterable in some way, such as regular expressions.
Second, interact from other software. Head to r-catmaid https://natverse.org/rcatmaid/ (part of the #natverse suite by Philipp Schlegel @uni_matrix, Alex Bates and others) for an R-based solution from the Jefferis lab at the #MRCLMB. Includes tools such as #NBLAST for anatomical comparisons of neurons (see paper by Marta Costa et al. 2016 https://www.sciencedirect.com/science/article/pii/S0896627316302653 ).
If R is not your favourite, then how about #python: the #navis package, again by the prolific @uni_matrix, makes it trivial, and works also within #Blender too for fancy 3D renderings and animations. An earlier, simpler version was #catpy by @csdashm https://github.com/ceesem/catpy , who also has examples on access from #matlab.
Third, directly from a #psql prompt. As in, why not? #SQL is quite a straightforward language. Of course, you'll need privileged access to the server, so this one is only for insiders. Similarly privileged is from an #ipython prompt initialized via #django from the command line, with the entire server-side API at your disposal for queries.
Fourth, and one of my favourites: from the #javascript console in the browser itself. There are a handful of examples here https://github.com/catmaid/CATMAID/wiki/Scripting but the possibilities are huge. Key utilities are the "fetchSkeletons" macro-like javascript function https://github.com/catmaid/CATMAID/wiki/Scripting#count-the-number-of-presynaptic-sites-and-the-number-of-presynaptic-connectors-on-an-axon and the NeuronNameService.getInstance().getName(<skeleton_id>) function.
Notice every #CATMAID server has its /apis/, e.g., at https://l1em.catmaid.virtualflybrain.org/apis/ will list all GET or REST server access points. Reach to them as you please. See the documentation: https://catmaid.readthedocs.io/en/stable/api.html
In short: the data is there for you to reach out to, interactively or programmatically, and any fine mixture of the two as you see fit.
#Drosophila #natverse #MRCLMB #django #javascript #VirtualFlyBrain #Platynereis #NBLAST #python #navis #blender #catpy #matlab #psql #sql #ipython #catmaid
Albert Cardona · @albertcardona
697 followers · 557 posts · Server qoto.orgAbove, my #introduction of interests. Here, who I am, what I do: a neuroscientist at the #MRCLMB and University of Cambridge, UK, studying the neural circuit basis of behavior, originally in #Drosophila but now also in #cephalopods (#pygmysquid #Idiosepius), the lancelet #Amphioxus and other animals. Our main approach: whole brain #connectomics with #vEM (volume electron microscopy) as the basis for computational modeling to guide neuronal activity perturbation and monitoring experiments with #optogenetics and #electrophysiology (#ephys for short).
Once upon a time I founded the #ImageJ -based #TrakEM2 software for image registration and neuronal arbor reconstruction and annotation, which spurred founding the #FijiSc (https://fiji.sc) image processing software, and later the #CATMAID web-based software for #connectomics.
Always open to inquires from prospective students and postdocs, and collaborations.
#introduction #vem #optogenetics #electrophysiology #ephys #imagej #TrakEM2 #MRCLMB #Drosophila #cephalopods #pygmysquid #Idiosepius #Amphioxus #connectomics #FijiSc #catmaid