Marc Robinson-Rechavi · @marcrr
1014 followers · 1969 posts · Server ecoevo.socialWelcome @bastien_boussau to Mastodon #MolecularEvolution #bioinformatics #ComputationalBiology
#MolecularEvolution #bioinformatics #computationalbiology
Benoit Nabholz · @benoitevol
336 followers · 267 posts · Server ecoevo.socialDo you think that using the term “evolutionary rate” for the ratio dN/dS is accurate or misleading?
#MolecularEvolution #EvolutionaryBiology #Evolution
Marc Robinson-Rechavi · @marcrr
934 followers · 1682 posts · Server ecoevo.socialGreat to see first keynote by an interdisciplinary paleobiologist: Phil Donoghue #smbe2023 #paleontology #MolecularEvolution
#smbe2023 #paleontology #MolecularEvolution
Marc Robinson-Rechavi · @marcrr
932 followers · 1678 posts · Server ecoevo.socialI'll be live-posting #SMBE2023 #SMBE23, please mute me for 4 days if you don't want lots of #MolecularEvolution in your timeline.
#smbe2023 #SMBE23 #MolecularEvolution
Marc Robinson-Rechavi · @marcrr
902 followers · 1566 posts · Server ecoevo.socialAnnouncing #SIB Comparative genomics course, taught by Christophe Dessimoz, Natasha Glover, @rmwaterhouse and myself, 30 August - 01 September in Lausanne #UNIL
• Introduction to molecular evolution & phylogenetics
• Orthology-focused arthropod comparative genomics
• Duplication of genes and genomes, expression
with #SIB resources #OMA, #Orthodb #BUSCO and @bgeedb #ortholog #orthologs #paralog #duplication #MolecularEvolution #Genomics #bioinformatics #training @P_Palagi https://www.sib.swiss/training/course/20230830_COMGE
#SIB #UNIL #oma #orthodb #busco #ortholog #orthologs #paralog #duplication #MolecularEvolution #Genomics #bioinformatics #training
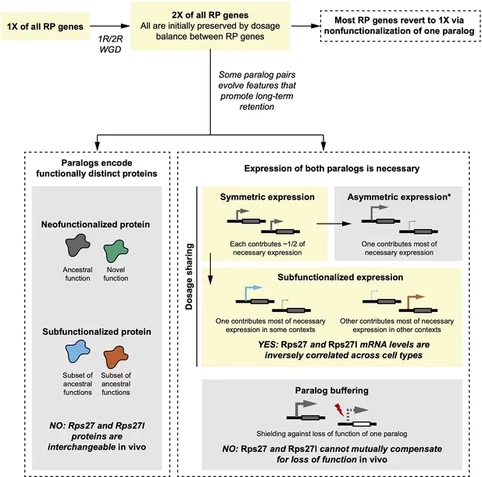
Stephen Matheson🌵🌲 · @sfmatheson
619 followers · 419 posts · Server fediscience.orgRps27 and Rps27l proteins are functionally interchangeable...
have been evolutionarily retained because the divergence of their expression patterns has resulted in both genes becoming necessary for achieving adequate total expression of the RP across cell types.
#MolecularEvolution
#Subfunctionalization
#geneduplication
#evolution
#evolution #geneduplication #subfunctionalization #MolecularEvolution
Paul Marrow 🇪🇺 · @evopma
420 followers · 639 posts · Server ecoevo.socialIn #Edinburgh for 50th anniversary of #rum #reddeerproject earlier this week. A very impressive and on-going study of #ecology #evolution #lifehistory #molecularevolution of Cervus elaphus L.. Many more eminent attendees than myself (only some found here @MarcoFB). My knowledge of the project was updated a lot.
#Edinburgh #rum #reddeerproject #ecology #Evolution #lifehistory #MolecularEvolution
Joanna Masel · @JoannaMasel
1061 followers · 417 posts · Server ecoevo.socialOur preprint describes how #EffectivePopulationSize affects #AminoAcid usage. https://www.biorxiv.org/content/10.1101/2023.02.01.526552v2. Within highly exchangeable pairs of amino acids, high Ne species are able to prefer #arginine over #lysine, and #valine over #isoleucine. This matches #thermophile preferences, as expected from theories of marginal protein stability at mutation-selection-drift balance. 1/6
@hanonmcshea #NearlyNeutralTheory #MolecularEvolution #EvolgenPaper
#effectivepopulationsize #aminoAcid #arginine #lysine #valine #isoleucine #thermophile #nearlyneutraltheory #MolecularEvolution #evolgenpaper
Marc Robinson-Rechavi · @marcrr
854 followers · 1302 posts · Server ecoevo.socialMasatoshi Nei, one of the founders of #MolecularEvolution, has died 😢. He was PhD or postdoc advisor to an amazing array of field leaders.
From: @JJ_Emerson
https://ecoevo.social/@JJ_Emerson/110396840914044756
Marc Robinson-Rechavi · @marcrr
829 followers · 1060 posts · Server ecoevo.socialLooks cool: Genomic and transcriptomic analyses support a silk gland origin of spider venom glands https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-023-01581-7 #EvoDevo #MolecularEvolution #Spider #venom
#evodevo #MolecularEvolution #spider #venom
Marc Robinson-Rechavi · @marcrr
825 followers · 1031 posts · Server ecoevo.socialJoanna Masel · @JoannaMasel
1030 followers · 340 posts · Server ecoevo.socialOur latest #preprint develops a new metric of #CodonAdaptation (not just #CodonBias) to measure the kind of #EffectivePopulationSize that matters to #NearlyNeutralTheory, and uses it to discover that #NaturalSelection prefers high #IntrinsicStructuralDisorder https://www.biorxiv.org/content/10.1101/2023.03.02.530449v1 1/6
#MolecularEvolution #PopulationGenetics #EvolgenPaper
#preprint #codonadaptation #codonbias #effectivepopulationsize #nearlyneutraltheory #naturalselection #IntrinsicStructuralDisorder #MolecularEvolution #populationgenetics #evolgenpaper
Marc Robinson-Rechavi · @marcrr
801 followers · 887 posts · Server ecoevo.social#Matreex can also be used for research into gene family evolution, providing a rapid and efficient overview of the gene family over many species. Visualization of intraflagellar transport gene families allowed us to discover complete loss in the #mixozoan Thelohanellus kitauei. #MolecularEvolution
#matreex #mixozoan #MolecularEvolution
Marc Robinson-Rechavi · @marcrr
801 followers · 886 posts · Server ecoevo.social#Matreex can be used for teaching, as here with the classical example of opsin evolution: notice the parallele loss of blue- and green-sensitive opsins in snakes and mammals, the duplications in frugivorous old-world primates, and the duplications and losses in fishes according to habitat. #opsin #MolecularEvolution
#matreex #opsin #MolecularEvolution
Joanna Masel · @JoannaMasel
997 followers · 314 posts · Server ecoevo.socialOur latest #preprint https://www.biorxiv.org/content/10.1101/2022.01.11.475913v2 @jdmatheson calls for significant change to the theory of #BackgroundSelection, a phenomenon by which some genomic segments of some individuals don’t count toward an #EffectivePopulationSize because they contain too many deleterious mutations. 1/5
#MolecularEvolution #PopulationGenetics #MutationLoad #EvolgenPaper
#preprint #backgroundselection #effectivepopulationsize #MolecularEvolution #populationgenetics #mutationload #evolgenpaper
Marc Robinson-Rechavi · @marcrr
779 followers · 734 posts · Server ecoevo.socialVer nice work by Martín-Zamora et al showing the role of #heterochrony of gene expression in the evolution of larva in bilaterians #Annelid #EvoDevo #MolecularEvolution https://doi.org/10.1038/s41586-022-05636-7
#heterochrony #annelid #evodevo #MolecularEvolution
Marc Robinson-Rechavi · @marcrr
777 followers · 702 posts · Server ecoevo.socialJust found the list of symposia of #SMBE23 #SMBE2023 online! Including "Integrating fossils and molecules to reconstruct the evolution of life in deep time" with the @moultdb team and many other cool topics. #MolecularEvolution
http://www.smbe.org/Portals/0/2023%20Meeting/SMBE2023-Symposia%2019%20DEC%202022.pdf
#SMBE23 #smbe2023 #MolecularEvolution
Marc Robinson-Rechavi · @marcrr
775 followers · 693 posts · Server ecoevo.socialAmazing seminar @dee_unil today by Magdalena Bohutínská (MajdaHolcova on Twitter). A researcher and a research program to follow! #ConvergentEvolution #MolecularEvolution #GenomeDuplication #PlantEvolution
https://ecoevo.social/@dee_unil/109699673384041182
#ConvergentEvolution #MolecularEvolution #genomeduplication #PlantEvolution
Marc Robinson-Rechavi · @marcrr
750 followers · 639 posts · Server ecoevo.socialCool concept by Fukushima & Pollock: measure selective convergent evolution by contrasting convergence (instead of rates) at non synonymous vs synonymous levels. #MolecularEvolution #NaturalSelection
https://doi.org/10.1038/s41559-022-01932-7
#MolecularEvolution #naturalselection
Marc Robinson-Rechavi · @marcrr
749 followers · 637 posts · Server ecoevo.socialJust read "Random genetic drift sets an upper limit on mRNA splicing accuracy in metazoans" by Benitiere et al @duret_lbbe, very nice and careful argument that most alternative splicing in e.g. vertebrates is non functional and accumulates neutrally. https://doi.org/10.1101/2022.12.09.519597 #MolecularEvolution #AlternativeSplicing #Neutralism
#MolecularEvolution #alternativesplicing #neutralism