Yann :python: · @nobodyinperson
101 followers · 343 posts · Server fosstodon.org@roadskater Compressed #CSV is (in my case) actually smaller than #NetCDF4. Trade IO speed for size and data approachability.
Yann :python: · @nobodyinperson
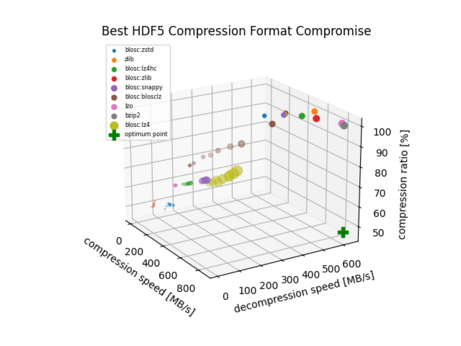
86 followers · 316 posts · Server fosstodon.org@thfriedrich I benchmarked the different compression algorithms in #HDF5 once if you're interested: https://gitlab.com/-/snippets/2043808
With the metric I use there (distance to optimum 'fast and small'), blosc:lz4 is the best compromise.
I still hit a wall with #HDF5 at some point though, I guess the compression prevented something from being done, I don't remember...
Also, #NetCDF4 is a subtype of #HDF5 so you'll feel familiar.
Thomas Friedrich · @thfriedrich
47 followers · 84 posts · Server fosstodon.org@nobodyinperson thanks for the insight. 🙂 I’ll have a look at #MQTT and #NetCDF4 which are both new to me. Working in science, so far I used mostly #hdf5 for larger data, which I think has a + for #openData & #openScience since there’s easy interfaces for most programming languages. Compression is decent I believe.
#mqtt #NetCDF4 #hdf5 #opendata #openscience
Yann :python: · @nobodyinperson
86 followers · 316 posts · Server fosstodon.org@thfriedrich Also, I've had problems with #NetCDF4 bindings not being thread-safe, so I couldn't parallelize operations very well. With compressed CSV, just throw threads (or processes) onto the problem. Files just work, no weird library in between 🙂
Yann :python: · @nobodyinperson
86 followers · 316 posts · Server fosstodon.org@thfriedrich Multidimensional would definitely be #NetCDF4, but if you are on a multidimensional scale with model-size outputs, then that's a different task than handling measurement device timeseries (what I was referring to).