Benedikt Mandl · @mandl
115 followers · 206 posts · Server mstdn.scienceRT @HartungIngo
The mystery why #BRD4 is so easily degradable by various #PROTACs seems to be solved: because it doesn’t need much to glue BRD4 to the E3 ligase #DCAF16. Stunning. I need to free up time today to read the 2 new papers @biorxivpreprint following the recent AbbVie paper @ACSBioMed!
CRAIG M CREWS · @crews
946 followers · 1224 posts · Server mstdn.sciencehttps://www.nature.com/articles/s41589-022-01218-w
Nice addition to the #targetedproteindegradation literature but I wouldn't describe it as "a new strategy of targeted protein degradation through direct substrate recruitment to the 26S proteasome" since Janse and Church first described this approach in 2004. https://pubmed.ncbi.nlm.nih.gov/15039430/ #PROTACs
#targetedproteindegradation #PROTACs
Adam M Gilbert · @adammgilbert
94 followers · 47 posts · Server mstdn.scienceRT @HartungIngo@twitter.com
Additional evidence about the likely structure of #IRAK4imid KT-413 @KymeraTx@twitter.com: a new patent application describes deuterated analogs of the compound shown in my previous post. Also in line with teachings from recent Kymera lectures on how to achieve po exposure. #PROTACs #TPD https://twitter.com/HartungIngo/status/1495714108712366081
🐦🔗: https://twitter.com/HartungIngo/status/1594638195471355907
Adam M Gilbert · @adammgilbert
48 followers · 28 posts · Server mstdn.scienceRT @HartungIngo@twitter.com
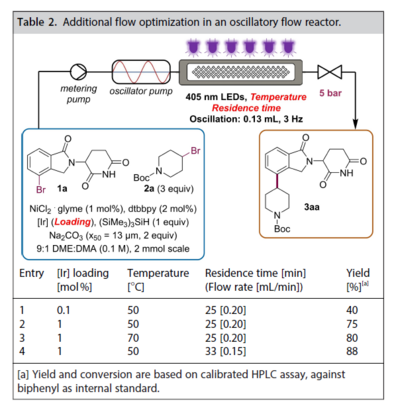
Ni-catalyzed photoredox couplings in an oscillatory flow reactor (to allow for handling of slurries) for accessing versatile CRBN-engaging building blocks for #PROTACs. From our (@merckgroup@twitter.com) collaboration with @KappeLab@twitter.com @UniGraz@twitter.com. @ChemCatChem@twitter.com https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cctc.202201184
🐦🔗: https://twitter.com/HartungIngo/status/1591417177479196672
CRAIG M CREWS · @crews
350 followers · 142 posts · Server mstdn.scienceI'd like to join in on some conversations so here are some #hashtags #TPD #targetedproteindegradation #PROTAC #PROTACs #degraders #chemicalbiology #biochem #cellbiology #biotech #biopharma #ligase #ubiquitin #proteasome #ligand #labhumor #chemistry #biology #proteome #proteins #drug #pharma #startup
#hashtags #TPD #targetedproteindegradation #PROTAC #PROTACs #degraders #ChemicalBiology #biochem #cellbiology #biotech #biopharma #ligase #Ubiquitin #proteasome #ligand #labhumor #chemistry #biology #proteome #proteins #drug #pharma #startup