Phil Brailey-Jones (he/him) · @Pretzel
85 followers · 233 posts · Server ecoevo.socialRT @SierraScientist@twitter.com
Still thinking about Denmark and #PlantMicrobeInteractions2022 Truly a conference of a lifetime!
🐦🔗: https://twitter.com/SierraScientist/status/1595449833904812032
Akos T Kovacs · @EvolvedBiofilm
223 followers · 330 posts · Server mstdn.scienceThis week, BIE group was at two conferences in 🇩🇰: @neftalyl and @RStannius presented their @cemist_dtu projects at the DMS 2022 congress, while @Xinming_Xu, @AdelePioppi, and @CajaDinesen discussed their CSC and #NNF_INTERACT projects at #PlantMicrobeInteractions2022
#NNF_INTERACT #PlantMicrobeInteractions2022
Akos T Kovacs · @EvolvedBiofilm
175 followers · 291 posts · Server mstdn.scienceGreat table companionship yesterday night at #PlantMicrobeInteractions2022 about wine, photography, science careers, and life stories…
Thank you, @SessitschAngela, Jos Raaijmakers, @EAlexandersson (#WineExpert), @RameshVetukuri, and @TonniGrube
#PlantMicrobeInteractions2022 #wineexpert
Akos T Kovacs · @EvolvedBiofilm
175 followers · 281 posts · Server mstdn.scienceAkos T Kovacs · @EvolvedBiofilm
175 followers · 281 posts · Server mstdn.scienceRT @ScienceCluster
Continuing the session on methods and #technologies for #plant #microbiome interrogation with @Manuel_Kleiner 🌱
The talk was on understanding key microbial functions related to #plant-microbe interactions #metaproteomics 🪴
#PlantMicrobeInteractions2022
@novonordiskfond
#technologies #plant #microbiome #metaproteomics #PlantMicrobeInteractions2022
Akos T Kovacs · @EvolvedBiofilm
175 followers · 281 posts · Server mstdn.scienceRT @ScienceCluster
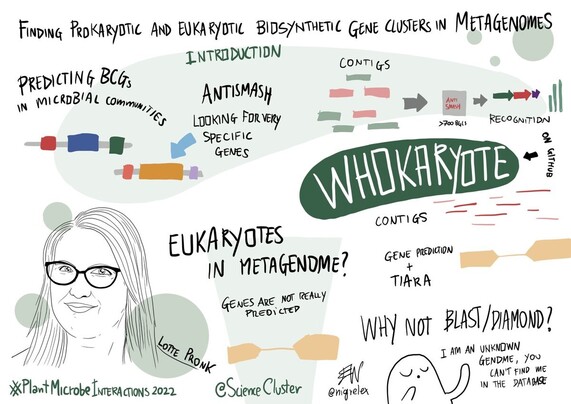
How to find #prokaryotic and #eukaryotic biosynthetic #gene clusters in #metagenomes? 🤔
@PronkLotte, the last speaker of #PlantMicrobeInteractions2022 is telling us more about it! 🦠
#science #plant #whokaryote
@novonordiskfond
#prokaryotic #eukaryotic #gene #metagenomes #PlantMicrobeInteractions2022 #Science #plant #whokaryote
Akos T Kovacs · @EvolvedBiofilm
169 followers · 272 posts · Server mstdn.scienceAmy Grunden (from @NCState) concludes the scientific part of #PlantMicrobeInteractions2022
That’s it, Folks!
#SlaapLekker
#PlantMicrobeInteractions2022 #SlaapLekker
Akos T Kovacs · @EvolvedBiofilm
169 followers · 270 posts · Server mstdn.scienceLotte highlights that indeed, they found new, fungal BGCs in the samples from @VCarryOn1 et al story above
➡️ application on metagenomes for visualizing eukaryotic BGCs
Akos T Kovacs · @EvolvedBiofilm
169 followers · 269 posts · Server mstdn.scienceAlso, need to separate pro- and eukaryote metagenome
Lotte introduces “Whokaryotes”, published in Microbial Genomics @MicrobioSoc 👇🏼
https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000823
Akos T Kovacs · @EvolvedBiofilm
169 followers · 269 posts · Server mstdn.scienceHowever, do we miss the eukaryotic share of BGCs in metagenomes❓
Most tools focus on prokaryotes ▶️ need to adapt pipelines for eukaryotes, e.g. introns in eukaryotes, different gene density
Akos T Kovacs · @EvolvedBiofilm
169 followers · 267 posts · Server mstdn.scienceLotte highlights antiSMASH and its usefulness to identify BGCs, even in metagenomes
Example used here: @VCarryOn1 et al in @ScienceMagazine on how to identify BGCs in metagenomes and meta transcriptomes ➡️over 700 BGCs
https://www.science.org/doi/10.1126/science.aaw9285
Akos T Kovacs · @EvolvedBiofilm
169 followers · 266 posts · Server mstdn.scienceLotte Pronk @PronkLotte (from @marnixmedema group at @WUR) on finding prokaryotic and eukaryotic biosynthetic gene clusters in metagenomes
#PlantMicrobeInteractions2022
Akos T Kovacs · @EvolvedBiofilm
168 followers · 264 posts · Server mstdn.scienceManuel on Roberto Kolter’s 7 members SynCom (assembled based on maize endophytes) in vitro and on plant proteomics, including general response of certain SynCom members to plant root colonization
Akos T Kovacs · @EvolvedBiofilm
168 followers · 263 posts · Server mstdn.scienceManuel describes their very recent study on plant microbiome proteome (note: lots of plant proteins as well that you can analyze)
#PlantMicrobeInteractions2022
---
RT @jmharris777
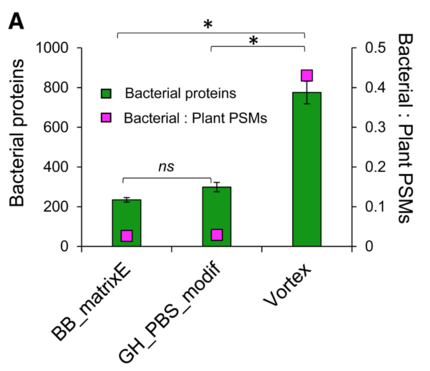
Evaluation of Protein Extraction Methods for Metaproteomic Analyses of Root-Associated Microbes - valuable analysis by Fernanda Salvato et al https://apsjournals.apsnet.org/doi/10.1094/MPMI-05-22-0116-TA #OpenAccess
https://twitter.com/jmharris777/status/1591603215430479872
#PlantMicrobeInteractions2022 #openaccess
Akos T Kovacs · @EvolvedBiofilm
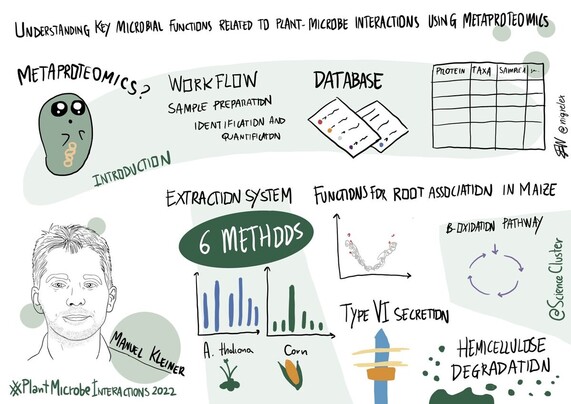
168 followers · 263 posts · Server mstdn.scienceManuel microbes in a microbiome, but what are they doing? 🟰 metaproteomics‼️
However, cell lysis is an important step in the pipeline
See @NatureComms 👇🏼
https://www.nature.com/articles/s41467-017-01544-x
Akos T Kovacs · @EvolvedBiofilm
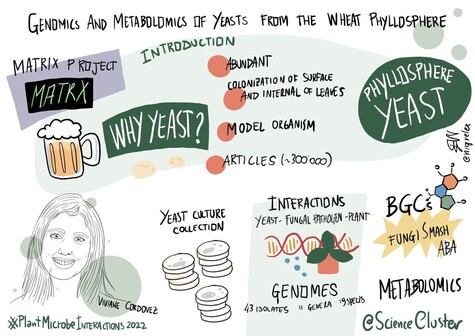
168 followers · 263 posts · Server mstdn.scienceRT @ScienceCluster
Have you ever wondered about the #genomics and #metabolomics of #yeasts from the wheat #phyllosphere? 🌾
Here is @VivianeCordovez speaking about it at #PlantMicrobeInteractions2022! 🌱
#conference #science
@novonordiskfond
#genomics #metabolomics #yeasts #phyllosphere #PlantMicrobeInteractions2022 #conference #Science
Akos T Kovacs · @EvolvedBiofilm
168 followers · 260 posts · Server mstdn.scienceManuel Kleiner @Manuel_Kleiner (from @NCState @NCStateCALS; #NNF_INTERACT #NNF_InRoot) on understanding key microbial functions related to plant-microbe interactions using metaproteomics
#NNF_INTERACT #NNF_InRoot #metaproteomics #PlantMicrobeInteractions2022
Akos T Kovacs · @EvolvedBiofilm
168 followers · 259 posts · Server mstdn.scienceRuben currently developing synthetic microbial ecosystems - turbidostat with OD controlled addition of carbon free medium
22 member SynCom for 125 days of closed systems▶️different types of interactions▶️is there evolution? [unpublished, but cool‼️]
#PlantMicrobeInteractions2022
Akos T Kovacs · @EvolvedBiofilm
168 followers · 258 posts · Server mstdn.scienceRuben also highlights that there is a great taxonomic overlap between root and physphere microbiota ➡️ testing SynCom based on abundances in the microbiomes➡️metabolic currencies exchanged in synthetic phycospheres
Akos T Kovacs · @EvolvedBiofilm
168 followers · 257 posts · Server mstdn.scienceRuben on characterization of Chlamydomonas phycosphere
See their @NatureComms paper👇🏼
https://www.nature.com/articles/s41467-022-28055-8
During time: soil ASV decrease with Phycosphere ASV increase in phycosphere samples 🟰similarly in Arabidopsis roots