PastelBio · @pastelbio
178 followers · 2000 posts · Server mstdn.scienceRT @NCIJBSloan: New resource coming from #CPTAC! It includes #proteomic and post-translational modifications, whole genome and whole exome sequencing, miRNA and totalRNA sequencing, imaging, and clinical information for over 1,000 cancer patients across 10 tumor types. https://proteomics.cancer.gov/news_and_announcements/introducing-suite-pan-cancer-multi-omic-papers-cptac
PastelBio · @pastelbio
172 followers · 1836 posts · Server mstdn.scienceRT @NCIDataSci: Access, analyze, & store #proteomic data in the updated search engine, PepQuery2! The tool was designed to empower proteomics researchers with quick #DataAnalysis. See how you can access it: https://datascience.cancer.gov/news-events/news/search-engine-tool-targets-peptide-mass-spectrometry-proteomic-data-sets
PastelBio · @pastelbio
150 followers · 1320 posts · Server mstdn.scienceA @pastelbio favourite #proteomic post from the last couple of months …
Pastel BioScience on Twitter https://twitter.com/PastelBio/status/1669273550065225729
UniSpec: A Deep Learning Approach for Predicting Energy-Sensitive Peptide Tandem Mass Spectra and Generating Proteomics-Wide In-Silico Spectral Libraries | https://www.biorxiv.org/content/10.1101/2023.06.14.544947v1 #proteomics
PastelBio · @pastelbio
138 followers · 1216 posts · Server mstdn.scienceA @pastelbio favourite #proteomic post from the last couple of months …
Pastel BioScience on Twitter https://twitter.com/PastelBio/status/1666359345498722306
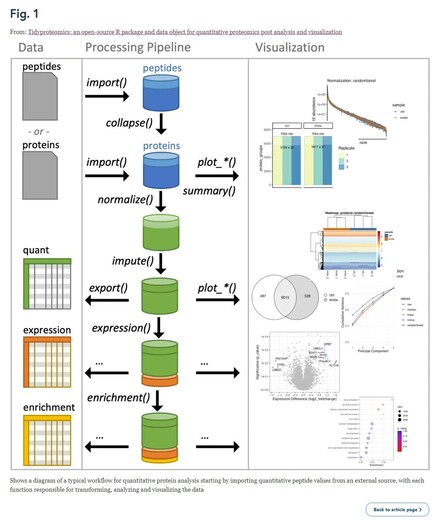
Tidyproteomics: an open-source R package and data object for quantitative proteomics post analysis and visualization | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-023-05360-7 #proteomics
PastelBio · @pastelbio
133 followers · 1134 posts · Server mstdn.scienceA @pastelbio favourite #proteomic post from the last couple of months …
Pastel BioScience on Twitter https://twitter.com/PastelBio/status/1669308792746520577
Evaluation of the Limit of Detection of Bacteria by Tandem Mass Spectrometry Proteotyping and Phylopeptidomics | https://www.mdpi.com/2076-2607/11/5/1170 #proteomics
PastelBio · @pastelbio
131 followers · 1080 posts · Server mstdn.scienceA @pastelbio favourite #proteomic post from the last couple of months …
Pastel BioScience on Twitter https://twitter.com/PastelBio/status/1648244984980680706
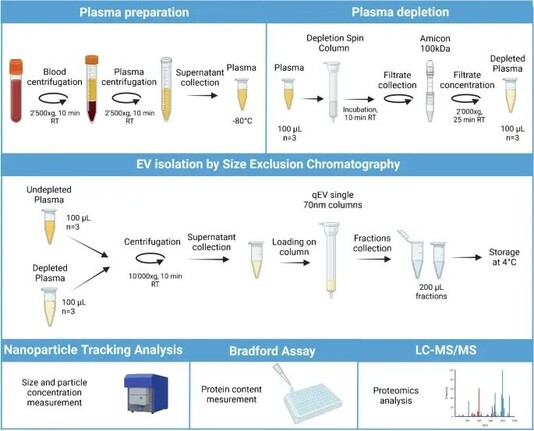
Depletion of abundant plasma proteins for extracellular vesicle proteome characterization: benefits and pitfalls | https://idp.springer.com/authorize?response_type=cookie&client_id=springerlink&redirect_uri=https%3A%2F%2Flink.springer.com%2Farticle%2F10.1007%2Fs00216-023-04684-w #proteomics
PastelBio · @pastelbio
119 followers · 466 posts · Server mstdn.scienceRT @isb_cgc: Proteomic Data Commons release V2.15 case, file, and quant data are now available in #BigQuery. Check out the new tables using our BigQuery Table Search tool at https://isb-cgc.appspot.com/bq_meta_search/ and filter Source by PDC.
#proteome #NCIProteomics #CancerResearch #proteomic https://twitter.com/isb_cgc/status/1622624155496480771/photo/1
#bigquery #proteome #nciproteomics #cancerresearch #Proteomic
jobRxiv · @jobRxiv
746 followers · 691 posts · Server mas.toMaster thesis/student researcher in Microbiology/Systems Biology/Proteomics @RalserLab
We're looking for a Master student/pre-doc to join our lab to study the #proteomic diversity of yeast isolates – join us in #Berlin!
@ChariteBerlin @ishengtsai
Cris Lapthorn · @makingions
184 followers · 200 posts · Server fosstodon.orgI thought electrospray was controversial! 😂
My rabbit hole today extends to trying to find robust and specific #MassSpec #proteomic and #metabolomics endpoints for apoptosis vs necrosis since the literature is littered with contradictory results for acetaminophen induced liver injury.😅
#massspec #Proteomic #metabolomics
FerRacimo · @FerRacimo
649 followers · 158 posts · Server fediscience.org*New #preprint alert* Patramanis et al. PaleoProPhyler: a #reproducible #pipeline for #phylogenetic inference using #ancient #proteins. 🦴
We developed a pipeline for aligning ancient #peptide data and performing #evolutionary analyses w/it. The pipeline can not only process #proteomic data, but also harness #genetic data in different formats (#CRAM, #BAM, #VCF) and translate it, so as to create reference panels for #phyloproteomics.
https://www.biorxiv.org/content/10.1101/2022.12.12.519721v1?med=mas
#ecoevo #evolution #ScienceMastodon #phyloproteomics #vcf #Bam #cram #genetic #Proteomic #evolutionary #peptide #proteins #ancient #phylogenetic #Pipeline #reproducible #preprint