PastelBio · @pastelbio
179 followers · 2119 posts · Server mstdn.scienceRT @byu_sam: #TeamMassSpec. question about tools for TMT quant. Which tools incorporate the MS1 intensity into the quantitation from MS2-based reporter ions?
I know that @nesvilab does this with Fragger. But does anyone else?
PastelBio · @pastelbio
179 followers · 2115 posts · Server mstdn.scienceRT @byu_sam: #TeamMassSpec. question about tools for TMT quant. Which tools incorporate the MS1 intensity into the quantitation from MS2-based reporter ions?
I know that @nesvilab does this with Fragger. But does anyone else?
PastelBio · @pastelbio
178 followers · 1965 posts · Server mstdn.scienceRT @DrLauraD22: Calling all #TeamMassSpec students interested in #proteomics. We have some excellent #PhD opportunities @WEHI_research proteomics facility. Please share! @FemalesInMS @PMV_Australia @LorneProteomics @hupo_org
#TeamMassSpec #proteomics #phd
PastelBio · @pastelbio
170 followers · 1788 posts · Server mstdn.scienceRT @byu_sam: After reading this, I am struck that all the negative examples (where CV is wonky) are because of terrible data quality, like letting in peptides with no FDR filter.
But is CV a bad metric if I am doing proper FDR filtering? #TeamMassSpec
PastelBio · @pastelbio
170 followers · 1783 posts · Server mstdn.scienceRT @: Assuming spray voltage is fine, what is the cause of the retrograde movement of the mobile along the emitter during ESI? #TeamMassSpec #Protoemics
Sarah Hancock · @sarahehancock
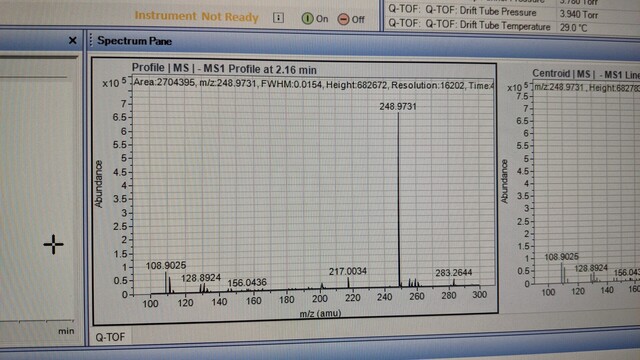
148 followers · 3 posts · Server mstdn.scienceJust in case anyone in #TeamMassSpec is curious, the likely culprit of my 248.9 m/z ion from this thread: https://mstdn.science/@sarahehancock/110818463525422168 is free cyanide reacting with the gold seals of the HPLC pump. I will have to try a few different brands of solvent to see if I can ameliorate this issue. More info also available here: https://community.agilent.com/technical/lcms/f/forum/5480/how-do-we-get-rid-of-identify-contamination-with-m-z-248-9737-negative-mode.
#TeamMassSpec #metabolomics #lipidomics #massspectrometry #massspec
PastelBio · @pastelbio
163 followers · 1657 posts · Server mstdn.scienceRT @: R package for annotating and visualizing high res MALDI proteoform/peptide data (and more). Congrats to David Degnan who has been driving this effort. #TeamMassSpec @J_ASMS
Sarah Hancock · @sarahehancock
147 followers · 2 posts · Server mstdn.scienceHey #TeamMassSpec, I'm having issues with a contaminant ion @ 248.9 m/z in -ve ion mode when running 90% ACN 10 mM AmOAc at pH 9. Found a thread on research gate suggesting it comes from the ACN but have tried two brands now with no joy. Has anyone else had this problem?
Research gate link: https://researchgate.net/post/What-is-this-contaminant-with-m-z-2489718
ACN brands tried: Fisher Sci Optima, Supelco LiChroSolv. LiChroSolv has a lot less of it than Fisher Sci.
#TeamMassSpec #metabolomics #lipidomics #massspectrometry #massspec
PastelBio · @pastelbio
164 followers · 1618 posts · Server mstdn.scienceRT @: Always good to see new #massspec tools available.Dear #proteomicscommunity and #TeamMassSpec, I am happy to share HowDirty, an R package that facilitates evaluating and reporting molecular contaminants in LC-MS experiments.
https://authorea.com/users/643346/articles/656759-howdirty-an-r-package-to-evaluate-molecular-contaminants-in-lc-ms-experiments…
#massspec #proteomicscommunity #TeamMassSpec
PastelBio · @pastelbio
164 followers · 1615 posts · Server mstdn.scienceRT @: *Job alert* An amazing opportunity is available to join our fabulous team @WEHI_research proteomics facility. Applications close 28 August. Please retweet! #TeamMassSpec
https://wehi.wd3.myworkdayjobs.com/en-US/WEHI/job/Proteomics-Research-Officer_JR2110-1…
PastelBio · @pastelbio
164 followers · 1600 posts · Server mstdn.scienceRT @: Folks! We have created a survey to better understand users' experience with OpenMS. The survey is short, and we would love to hear your opinions on what's good and bad! Please share your thoughts
#proteomics #metabolomics #lipidomics #TeamMassSpec #phd
PastelBio · @pastelbio
159 followers · 1556 posts · Server mstdn.scienceRT @: Deadline Sept 30th!
Call for papers @JProteomeRes Special Issue
"Women in #proteomics and #metabolomics"
https://axial.acs.org/biology-and-biological-chemistry/cfp-jprobs-women-in-proteomics-and-metabolomics…
@FemalesInMS #WomenInSTEM @AmerChemSociety #Teammassspec
#proteomics #metabolomics #womeninSTEM #TeamMassSpec
PastelBio · @pastelbio
158 followers · 1504 posts · Server mstdn.scienceRT @: Anyone have advice on how they normalize TMT channel data when the samples are from a pulldown? Normalization methods assume that peptide abundances should be broadly equal across samples...
#TeamMassSpec #massspec #proteomics
PastelBio · @pastelbio
155 followers · 1421 posts · Server mstdn.scienceRT @: Last reminder: everyone attending #BSPREUPA2023 check out your email to learn all about the YPIC activities! Join us for dinner on Sunday, workshops on Monday, breakfast on Tuesday or during any break at our booth! #TeamMassSpec #conference @UKBspr @EuPAProteomics
#bspreupa2023 #TeamMassSpec #conference
PastelBio · @pastelbio
153 followers · 1367 posts · Server mstdn.scienceRT @: Less than one week to go for #BSPREUPA2023
Check out the updated programme:
https://conferences.ncl.ac.uk/bspr-eupa2023/programme/…
@UKBspr @EU_YPIC @EuPAProteomics
#bspreupa2023 #TeamMassSpec #proteomics
PastelBio · @pastelbio
147 followers · 1312 posts · Server mstdn.scienceRT @: Announcing the 2023 @asmsnews Asilomar Conference on Computational Mass Spec.
Abstract submission, registration, lodging, and stipend application are all OPEN.
https://asms.org/conferences/asilomar-conference…
RT #TeamMassSpec @olgavitek @DavidFenyo
PastelBio · @pastelbio
147 followers · 1295 posts · Server mstdn.scienceRT @: Hey #TeamMassSpec - question re: low sample stage-tip fractionation. Anyone have any experience with it? We have some POROS 20 R2 packing. How much (~mg?) do you load into a 200µL tip?
#TeamMassSpec #massspec #proteomics
PastelBio · @pastelbio
147 followers · 1295 posts · Server mstdn.scienceRT @: #BSPREUPA2023 is just over 1.5 weeks away!
I'm getting more and more excited as the programme was just released! https://conferences.ncl.ac.uk/bspr-eupa2023/programme/…
#TeamMassSpec #proteomics @UKBspr @EU_YPIC @EuPAProteomics
#bspreupa2023 #TeamMassSpec #proteomics
Dr Nick Birse · @njbirse
38 followers · 8 posts · Server mstdn.scienceHi - trying Mastodon again, last tried it in December 2022.
So, I'm a pretty new Lecturer in Mass Spectrometry at Queen's University Belfast.
My BSc was Pharmaceutical Chemistry at Dundee, followed by an MSc in Instrumental Analytical Science at Robert Gordon, and a PhD (plus postdoc) in mass spectrometry at Queen's.
If you do/like any of that lot (analytical chemistry and toxicology, really) then I'd love to chat.
Dr Nick Birse · @njbirse
38 followers · 8 posts · Server mstdn.scienceWelcome back to my Mastodon feed.
I'm now a Lecturer in Mass Spectrometry at Queen's University Belfast, and can still be found working on all sorts of mass spectrometry for food and environmental samples.