Dr Emma Hodcroft :verified: · @firefoxx66
2841 followers · 123 posts · Server mstdn.scienceI had a truly brilliant time at #VGE22 in #Hinxton! So many fantastic posters, talks & people, I could have stayed much longer to have even more discussions!
Many thanks to @katrina_lythgoe@twitter.com @emcat1@twitter.com @christian_happi@twitter.com @KateGrabowski@twitter.com for such a fantastic conference! ❤️😁🧬🌳
Gage Moreno · @gage
43 followers · 14 posts · Server mstdn.scienceLeaving #VGE22 feeling refreshed and full of new ideas. It was a wonderful time meeting everyone. Can’t wait for May 2024 when we get to do it all again 🦠🤩
Dr Emma Hodcroft :verified: · @firefoxx66
2841 followers · 123 posts · Server mstdn.scienceRT @ifeanyi_omah@twitter.com
No time has phylogenetic been more important than now in epidemiology of infectious diseases. @firefoxx66@twitter.com providing insight on what is next in sequencing and phylogenetic and how to scale what we have learned during the pandemic to tackle other pathogens. #VGE22
🐦🔗: https://twitter.com/ifeanyi_omah/status/1591063131274764289
Gage Moreno · @gage
43 followers · 14 posts · Server mstdn.scienceFascinating detective work🕵️♀️ by @emcat1@twitter.com investigating non-A-E hepatitis in children. A tour de force combination of viral and host genomics, histopathology, and serology to pinpoint AAV2 as probably cause of infection. #VGE22
Dr Emma Hodcroft :verified: · @firefoxx66
2841 followers · 123 posts · Server mstdn.scienceRT @GageKMoreno@twitter.com
Genomic sequencing & phylogenetics can and needs to be implemented on commonly circulating viruses (RSV, influenza) rather than waiting for the next pandemic. It’s up to all of us to push the boundaries of generating this data. Great talk @firefoxx66@twitter.com! #VGE22
🐦🔗: https://twitter.com/GageKMoreno/status/1591016259608809472
Tim Downing · @downingtim
87 followers · 80 posts · Server genomic.socialAdeno-associated virus 2 #aav2 is the cause of unexpected #hepatitis in children understood via #metagenomics #VGE22 - Emma Thomson
#VGE22 #metagenomics #hepatitis #aav2
Gage Moreno · @gage
43 followers · 14 posts · Server mstdn.sciencePatrick Varilly giving a demo of how we can begin to generate MCMC phylogenetic trees in real time. Fully converged tree with results comparable to BEAST2 in as little as 8 minutes while running locally on his laptop @fathominfo@twitter.com @sabeti_lab@twitter.com #VGE22
Gage Moreno · @gage
43 followers · 14 posts · Server mstdn.scienceGenomic sequencing & phylogenetics can and needs to be implemented on commonly circulating viruses (RSV, influenza) rather than waiting for the next pandemic. It’s up to all of us to push the boundaries of generating this data. Great talk @firefoxx66@twitter.com! #VGE22
Dr Emma Hodcroft :verified: · @firefoxx66
2841 followers · 123 posts · Server mstdn.scienceRT @BortzGroup@twitter.com
Dr Emma Hodcroft @firefoxx66@twitter.com of @nextstrain@twitter.com leads off #VGE22 with an insightful look into the essential global role of virus genome sequencing in pandemic response, from #SARSCoV2 to the next pathogen. She highlights sharable resources: 1. Software, 2. People, 3. Data sharing.
🐦🔗: https://twitter.com/BortzGroup/status/1591007001114546176
Gage Moreno · @gage
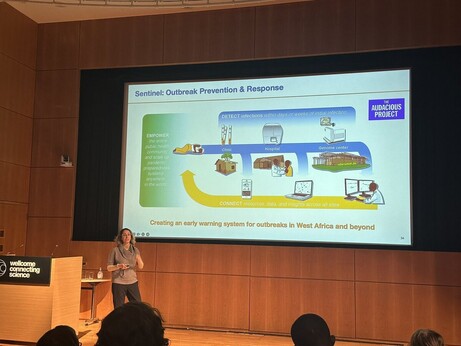
43 followers · 14 posts · Server mstdn.scienceA fantastic keynote from @PardisSabeti@twitter.com with lessons from past outbreaks and insights into what we can do to prevent the next pandemic. Key insights: normalizing surveillance infrastructure (i.e. Bluetooth tracking) in downtime rather than responsive implementation #VGE22
Tim Downing · @downingtim
87 followers · 80 posts · Server genomic.social#VGE22 talk by Jonathan Henney on vaccine prep. #flu A(H5) clade 2.3.4.4 may be severe if it becomes widespread, so create #vaccine reflecting avian-pig-human mosiacs from antigenically shifted #recombinants. #genomics
#genomics #recombinants #vaccine #flu #VGE22
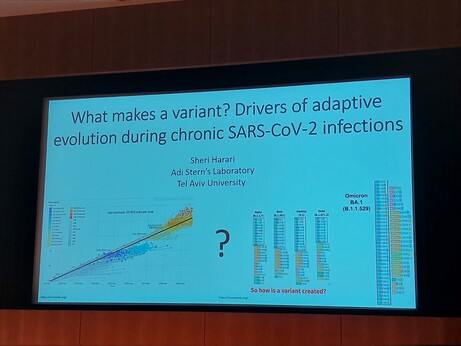
Tim Downing · @downingtim
87 followers · 80 posts · Server genomic.socialInteresting #chroic #virus #genomics work outlined by Tess Lambe - #hiv capsid region subject to weaker per site positive selection, with a higher syn than nonsyn site change rate in vivo during longterm infections compared to between patients #VGE22
#VGE22 #hiv #genomics #Virus #chroic
Gage Moreno · @gage
43 followers · 14 posts · Server mstdn.scienceInsights from @tanyagolubchik@twitter.com on #RSV 🦠 severity in children and infants pre-#SARSCoV2. Confections, especially H. influenzas, lead to needing more support from increased severity. Very important implications for the current RSV surge seen in young children. #VGE22
Tim Downing · @downingtim
87 followers · 80 posts · Server genomic.socialGreat talk on viral load correlates from #genome libraries (targeted #metagenomics) in #RSV and co-infecting viruses in children by Tanya Golubchik #VGE22
#VGE22 #rsv #metagenomics #genome
Gage Moreno · @gage
43 followers · 14 posts · Server mstdn.scienceRT @sabeti_lab@twitter.com
Sabeti Lab superstars @lakrasil@twitter.com @PetrosBrittany@twitter.com @GageKMoreno@twitter.com giving lightning talks at #VGE22
Gage Moreno · @gage
43 followers · 14 posts · Server mstdn.scienceOliver Pybus ends his talk saying we need to start pairing genomes with serology, demographics, and other metadata to maximize the value of genomic surveillance systems #VGE22
Dr Emma Hodcroft :verified: · @firefoxx66
2841 followers · 123 posts · Server mstdn.scienceNabil-Fareed Alikhan · @happykhan
661 followers · 255 posts · Server mstdn.scienceRT @K_G_Andersen@twitter.com
I gave a talk earlier today at #VGE22 on “The Proximal Origin and Evolution of SARS-CoV-2”.
Slides are here: https://andersen-lab.com/files/VGE_2022.pdf
Recorded talk (25mins) here: https://www.youtube.com/watch?v=2nZB07FV4Ug
🐦🔗: https://twitter.com/K_G_Andersen/status/1590407745324617729
Dr Emma Hodcroft :verified: · @firefoxx66
2841 followers · 123 posts · Server mstdn.scienceGreat talk by @LemeyLab@twitter.com at #VGE22 touching on the need for better understanding of #RSV.
Q&A highlighted how critical it is to have data analyses & sources that allow others to build upon them easily & openly - an ecosystem @nextstrain@twitter.com is proud to be part of!
Dr Emma Hodcroft :verified: · @firefoxx66
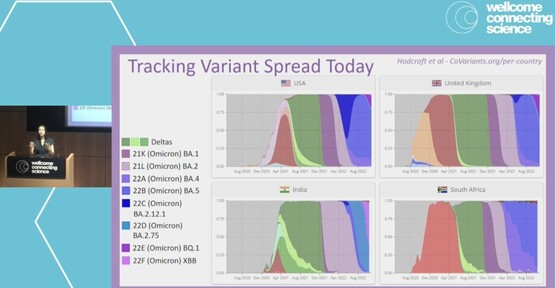
2841 followers · 123 posts · Server mstdn.scienceInteresting question to @Tuliodna@twitter.com at #VGE22 -- "what will happen with #SARSCoV2 after the world cup?"
With people coming together from all over the world, what might this mean for cases, variant spread, even new variants?
Indeed, worth keeping close surveillance on!