AnarchistSpaceDad · @freakazoid
1023 followers · 24912 posts · Server retro.socialWent to check on how #DeepMind did at #CASP15 and discovered that #AlphaFold did not even enter. Instead, most of the participants used AlphaFold 2 or some variant. So DeepMind has revolutionized protein folding prediction. With all the hype around #AI, why do real gains like this get ignored in favor of toys?
#deepmind #casp15 #alphafold #ai
Arne Elofsson · @arneelof
143 followers · 435 posts · Server fediscience.orgOn Wednesday Jan 11, at 17:00 (Swedish Time) we will have the first CASP zoom meeting. All information can be found at casp.bioinfo.se. Here is a short recap:
Mailinglist: casp-zoom@googlegroups.com
Zoom: https://stockholmuniversity.zoom.us/j/61626785695
The first meeting will be dedicated to discussing how to proceed with these meetings.
Arne Elofsson · @arneelof
91 followers · 296 posts · Server fediscience.orgMy take on the last year of Protein structure prediction up and including #CASP15 comments are welcome.
[2212.07702] Protein Structure Prediction until CASP15 https://arxiv.org/abs/2212.07702
Arne Elofsson · @arneelof
91 followers · 295 posts · Server fediscience.orgMy take on the last year of Protein structure prediction up and including #CASP15 comments are welcome.
[2212.07702] Protein Structure Prediction until CASP15 https://arxiv.org/abs/2212.07702
Arne Elofsson · @arneelof
88 followers · 288 posts · Server fediscience.org
Arne Elofsson · @arneelof
88 followers · 286 posts · Server fediscience.org#CASP15 in Nature is out now https://www.nature.com/articles/d41586-022-04438-1
Arne Elofsson · @arneelof
86 followers · 284 posts · Server fediscience.orgRT @dxbarradas@twitter.com
Last day of #CASP15 - discussions about what is an interface ? Very interesting 😃
🐦🔗: https://twitter.com/dxbarradas/status/1602572525480935424
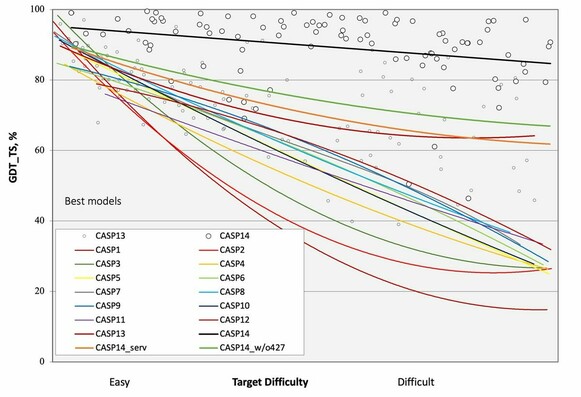
Albert Vilella · @albertvilella
345 followers · 199 posts · Server genomic.socialSlides of the different #CASP15 presentations available for download.
Slide below shows how #CASP14 (black line) was truly the renaissance moment of protein folding
RT @sokrypton@twitter.com
#CASP15 slides from days 1-3 released!
https://predictioncenter.org/casp15/doc/presentations/
🐦🔗: https://twitter.com/sokrypton/status/1602308662260908032
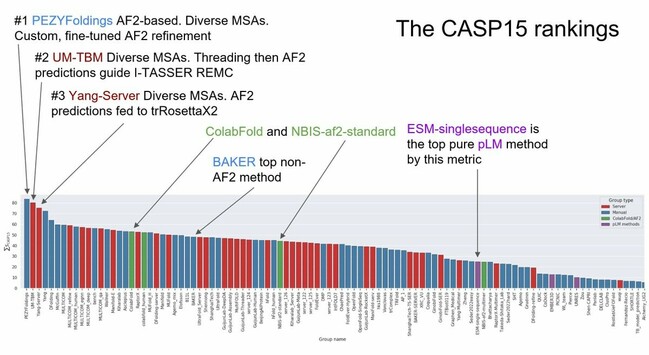
Albert Vilella · @albertvilella
345 followers · 199 posts · Server genomic.socialMore notes from the #CASP15 competition. Other resources here: https://www.aditiashenoy.com/posts/casp15/day2/
RT @AlbertVilella@twitter.com
Some initial results from the #CASP15 competition and we see that #Alphafold2 has become a fertile ground for experimentation by several research groups around the world. Facebook's ESM protein language models (pLMs) are the top non-MSA based methods.
🐦🔗: https://twitter.com/AlbertVilella/status/1601909663309901824
Albert Vilella · @albertvilella
340 followers · 196 posts · Server genomic.socialAnother take-away from #CASP15 is that people are now starting to consider large-scale computational Protein Protein Interaction analyses (PPIs). These have been done experimentally for more than 30 years in a high-throughput manner, e.g. for yeast. https://www.aditiashenoy.com/posts/casp15/day2/
Albert Vilella · @albertvilella
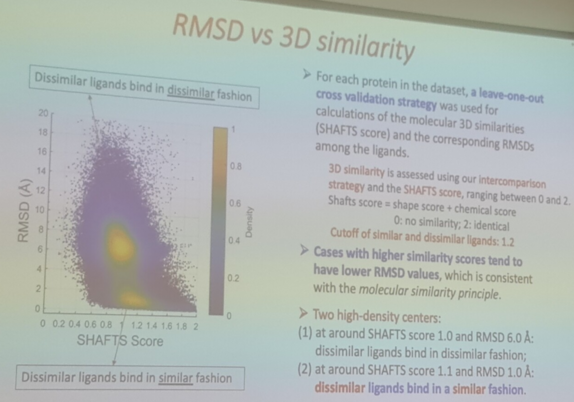
340 followers · 194 posts · Server genomic.socialAlbert Vilella · @albertvilella
340 followers · 193 posts · Server genomic.socialContinuing with #CASP15, now on the topic of ligands and their prediction on existing protein structures.
Arne Elofsson · @arneelof
82 followers · 277 posts · Server fediscience.orgArne Elofsson · @arneelof
82 followers · 277 posts · Server fediscience.orgRT @bjornwallner@twitter.com
@gabriielstuder@twitter.com QA assessor in #CASP15 reminds us about the length dependence of TMscore. Problematic for long sequences.
🐦🔗: https://twitter.com/bjornwallner/status/1602549513314570241
Arne Elofsson · @arneelof
82 followers · 273 posts · Server fediscience.orgArne Elofsson · @arneelof
82 followers · 272 posts · Server fediscience.orgArne Elofsson · @arneelof
82 followers · 272 posts · Server fediscience.orgRT @aditiashenoy@twitter.com
Takeaways from Day 3 at #CASP15
https://www.aditiashenoy.com/posts/casp15/day3/
🐦🔗: https://twitter.com/aditiashenoy/status/1602419271337910272
Arne Elofsson · @arneelof
82 followers · 267 posts · Server fediscience.orgThis is from discussion yesterday and will be discussed further today give me your thoughts
My suggestion is that we schedule monthly meetings at the last Monday in the month at 17:00 (5pm) Swedish time. This is (normally), 16:00, UK time, 11:00 am in New York, 8:00 am San Francisco, 22:00 in Beijing, I think.
Lucas Nivon · @lucas_nivon
128 followers · 244 posts · Server mas.toLots of content about #casp15 on the incipient #proteinengineering mastodon! I missed this exciting preprint using Bakerlab's RF Diffusion led by Susana Vazquez Torres to bind small helical peptides. Very hard design target with pre-DL Rosetta methods. https://www.biorxiv.org/content/10.1101/2022.12.10.519862v2
Lucas Nivon · @lucas_nivon
186 followers · 372 posts · Server mas.toLots of content about #casp15 on the incipient #proteinengineering mastodon! I missed this exciting preprint using Bakerlab's RF Diffusion led by Susana Vazquez Torres to bind small helical peptides. Very hard design target with pre-DL Rosetta methods. https://www.biorxiv.org/content/10.1101/2022.12.10.519862v2