Daniel Caron · @carondanielp
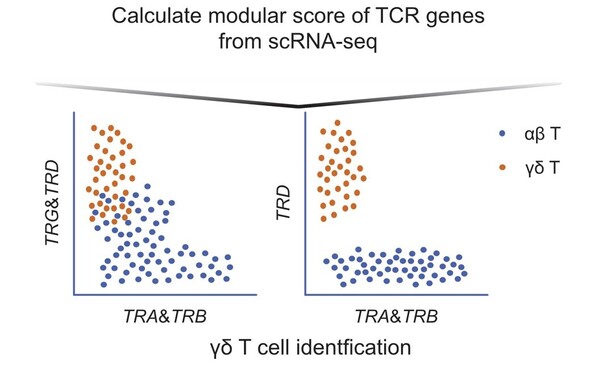
44 followers · 46 posts · Server genomic.socialCool computational method for separating γδ #Tcells by #rna #sequencing
Rather than relying on paired #TCRseq or #CITEseq, they calculate module scores of TCR constant and variable regions
#Tcells #RNA #sequencing #tcrseq #citeseq
Daniel Caron · @carondanielp
32 followers · 37 posts · Server genomic.socialSo excited to share MMoCHi: a tool I've built for multi-modal cell type classification for CITE-seq data
The pre-print can be found here:
https://www.biorxiv.org/content/10.1101/2023.07.06.547944v1
And we've put it out on #GitHub so that you can try it out on your own data:
https://mmochi.readthedocs.io
#scRNAseq #scRNA #CITEseq #sequencing #analysis #ColumbiaUniversity #immunology
#github #scrnaseq #scRNA #citeseq #sequencing #analysis #columbiauniversity #immunology
Haojia Wu · @haojiawu
149 followers · 47 posts · Server mastodon.haojia-wu.comA new multiomics study from Satija Rahul's lab identified a unique population of CD8+ T cells after COVID-19 vaccination. These cells are the SARS-CoV-2 antigen specific and capable of rapid clonal expansion. The relative frequency and differentiation of these cells can predict clinical outcomes for COVID-19 patients. #covid #multiomics #scRNAseq #scATACseq #CITEseq #ASAPseq #TCR
https://www.biorxiv.org/content/10.1101/2023.01.24.525203v1
#covid #multiomics #scrnaseq #scatacseq #citeseq #asapseq #tcr
Haojia Wu · @haojiawu
26 followers · 11 posts · Server mastodon.haojia-wu.comProx-seq for simultaneous measurement of proteins, protein complexes and mRNAs in thousands of single cells! With this multi-modal profiling technique, the authors were able to directly measure the protein complexes on naïve CD8+ T cells and protein interactions during TLR4 activation in macrophages. #singlecell #multimodal #newtech #CITEseq #REAPseq
https://www.nature.com/articles/s41592-022-01684-z
#singlecell #multimodal #newtech #citeseq #reapseq