jwrth · @jwrth
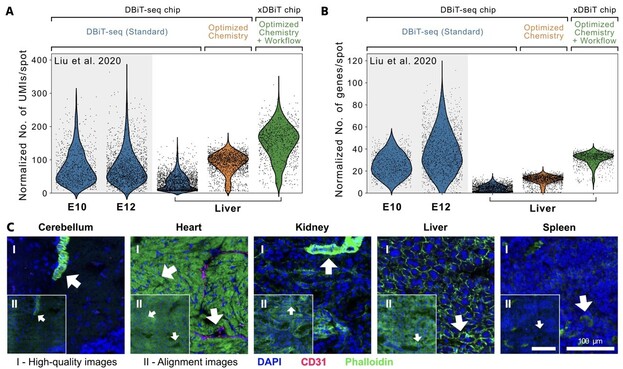
14 followers · 6 posts · Server genomic.socialImprovements in workflow and #chemistry lead to increased numbers of reads and counts per spot compared to the original #DBiTseq method. An extended #imaging workflow prevents the destruction of image features. 🧬🔬 2/6
jwrth · @jwrth
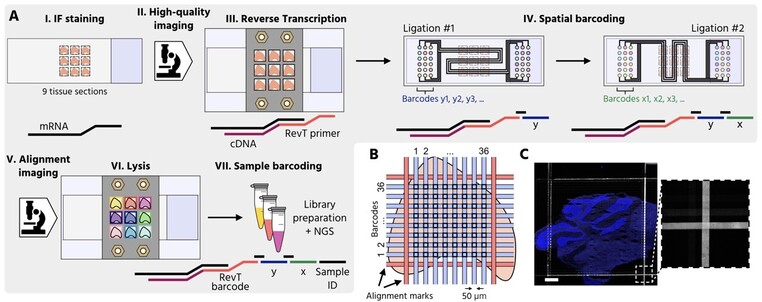
14 followers · 5 posts · Server genomic.socialI am excited to present multiplexed Deterministic Barcoding in Tissue (xDBiT)🎊. As an extension of #DBiTseq it allows the generation of #spatial #transcriptomics datasets of nine fixed tissue sections in parallel. Paper finally out on @NatComms @Nature 📜 https://www.nature.com/articles/s41467-023-37111-w 1/6
#dbitseq #Spatial #Transcriptomics
KelvinTuong · @KelvinTuong
42 followers · 80 posts · Server genomic.socialRT @Kelvyinzinho
Delighted to share our latest collab project led by @johwirth with @Meier_Lab. 🤩 #spatial #transcriptomic #DBiTseq
Check it out👇🏼
Spatial transcriptomics using multiplexed deterministic barcoding in tissue | Nature Communications https://www.nature.com/articles/s41467-023-37111-w
#Spatial #transcriptomic #dbitseq