PastelBio · @pastelbio
153 followers · 1388 posts · Server mstdn.scienceRT @: PTM-containing peptides often fragment in unexpected ways, hindering their identification. Here we describe a workflow in #FragPipe to find diagnostic spectral features for any PTM, illustrated using chemoproteomics, RNA-xlinks, glyco and ADP-ribo examples
Ben Neely · @neely
265 followers · 320 posts · Server fediscience.orgWant to thank everyone here for some stellar discussion on SAAVs and searching.
Follow up: is there a SAAV-only open search option? So open-search but constrain fitting delta mass unknowns to AAs only (no funky ptms). I am off to poke around in #FragPipe but figured the crowd might know better.
#teamproteome #teammassspec #fragpipe
PastelBio · @pastelbio
119 followers · 466 posts · Server mstdn.scienceRT @kalonji_08: Does anyone know how to Install #Fragpipe and all dependencies in Linux ? I’m having some #Java problem and I wouldn’t want to risk running it on my laptop. Please assist Thanks #massspec #Bioinformatics #proteomics
#fragpipe #Java #massspec #bioinformatics #proteomics
PastelBio · @pastelbio
119 followers · 466 posts · Server mstdn.scienceRT @nesvilab: Meet our #FragPipe team at #USHUPO23! We present our latest results on deep learning rescoring, HLA peptidomics, labile PTM searches, single cell proteomics (DIA,LFQ-MBR, TMT), plus two short courses. Find us, or contact me to schedule a meeting to discuss new collaborations etc.
Edu · @educhicano
115 followers · 144 posts · Server masto.esRT @TTCooper_PhD@twitter.com
Everyone in #proteomics and #massspectometry land. I have made a tutorial on how to handle multi-CV FAIMS DDA/DIA front to back using #python and #fragpipe. Room for improvement, but it works.
https://link.medium.com/OtfAGYWeLwb
🐦🔗: https://twitter.com/TTCooper_PhD/status/1616664991452565505
#proteomics #massspectometry #python #fragpipe
Fatih Demir · @kabalak
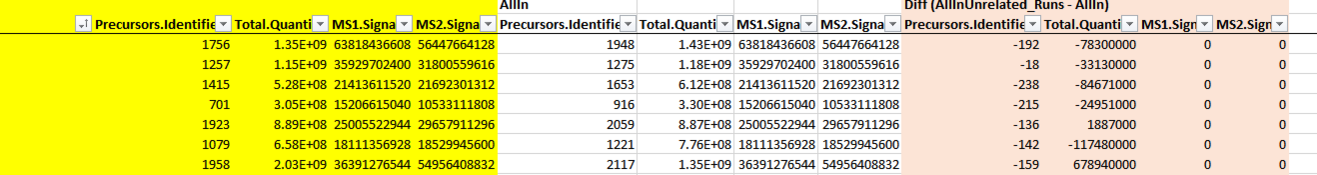
53 followers · 98 posts · Server mstdn.scienceA question for all the DiaNN experts here: I run DiaNN through FragPipe 19.1 and tried to see if there is a big deal in the "Unrelated runs" option in FragPipe for DiaNN. I totally understand the ID data differing, but not why the Total.Quantity if different for both run modi, though the MS1 and MS2 Quantities are identical?!? (in red the difference) #FragPipe #DiaNN
Edu · @educhicano
113 followers · 117 posts · Server masto.esRT @nesvilab@twitter.com
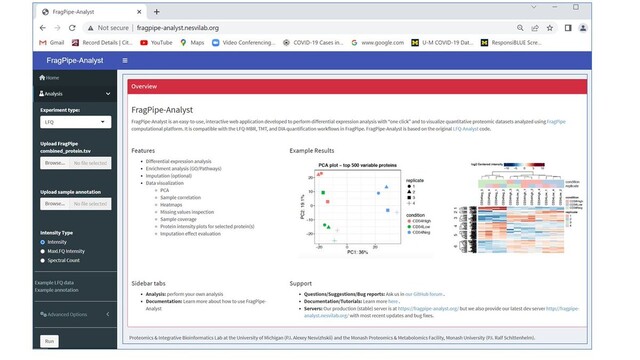
#FragPipe team, in collaboration with the Monash University @RalfSchittenhe1@twitter.com and extending their LFQ-Analyst, presents FragPipe-Analyst. Load FragPipe's TMT/ LFQ/DIA quant tables and get PCA, volcano plots, heatmaps, do GO/pathway enrichment analysis, all with just a few clicks!
Brian Hampton · @ionurchin
32 followers · 73 posts · Server mastodon.social#TeamMassSpec
I use #FragPipe developed by the Nesvizhskii lab for #proteomics data analysis. The latest update has a link to analyze the results using a version of #LFQAnlyst compatible with FragPipe output called #FragPipe-Analyst.
http://fragpipe-analyst.nesvilab.org/
This is a powerful R statistical analysis workflow that anybody can quickly use to get descriptive and differential abundance results directly from the FragPipe output. Thanks @nesvilab
#TeamMassSpec #fragpipe #proteomics #lfqanlyst