Alan · @alan
6 followers · 55 posts · Server piaille.frThinking about publishing some #experimental data in fluid dynamics in an open format that could be a standard, and trying not to reproduce this piece of art
https://xkcd.com/927/
#hdf5 looks interesting, or #netCDF but my data are not so standardized.
gauteh · @gauteh
24 followers · 45 posts · Server hachyderm.ioFixed a long standing bug in hidefix (https://github.com/gauteh/hidefix) where #hdf5 files with un-aligned chunking failed. 0.7.0 is out. Fast as ever compared to native hdf5 library. #rust
nialov · @nialov
3 followers · 37 posts · Server fosstodon.orgLooking for help with hdf5 and fortran packaging as a complete newbie: https://github.com/NixOS/nixpkgs/issues/248503
Markus Osterhoff · @mosterh1
87 followers · 499 posts · Server academiccloud.socialAnd after … a couple of days, here the #bugfix for my #memoryLeak
It turns out that using #HDF5 needs some cleanup routines – that's totally okay, but I have plenty of #Terabytes that worked without 🤔
#terabytes #hdf5 #memoryleak #bugfix
Markus Osterhoff · @mosterh1
81 followers · 486 posts · Server academiccloud.socialThe #HDF User Group will hold their annual European meeting on 19-21 September at #DESY, Hamburg, Germany.
The special topic of this meeting will be #HDF5 plugins and #data #compression, and the meeting is hosted in collaboration with the LEAPS-INNOV EU project. The HDF Group will give an update on the latest developments within HDF5 and present the roadmap for the future.
#compression #data #hdf5 #desy #hdf
Markus Osterhoff · @sci_photos
235 followers · 3309 posts · Server troet.cafeEin anderes Programm (nennen wir es kleinhirn) hat sinnvollere Werte angezeigt. Aber das erste (nennen wir es zeiger) soll zählen, die Werte an das grosshirn senden, wo SPEC die Zahlen dann abholt für die Justage.
Die Daten kommen als #LZ4-komprimierte #HDF5-Strom; zeiger entpackt und zählt, schreibt anschließend den originalen lz4-Blob in die Datei; kleinhirn auf einem anderen Rechner öffnet die Datei, zählt auch und zeigt das Bild an.
gauteh · @gauteh
18 followers · 26 posts · Server hachyderm.ioNew release of hidefix (https://github.com/gauteh/hidefix) updated to work with xarrays plugin registration. pip install hidefix should do the trick, conda package in the wait: https://github.com/conda-forge/staged-recipes/pull/21742 #hidefix #rust #hdf5 #netcdf
Christopher J Burke · @CuriousTerran
101 followers · 196 posts · Server universeodon.comWarrick Ball · @warrickball
75 followers · 347 posts · Server mas.toI vote for whatever will be most easily queried by user tools like astroquery (or the equivalent of Lightkurve, if one is made).
That said, I lean towards #HDF5. I wish I'd started using it more and sooner.
Mark Kittisopikul · @markkitti
251 followers · 114 posts · Server fosstodon.org#HDF5 1.14.0 builds are about to land in #condaforge and #msys2. Please review if interested.
gauteh · @gauteh
10 followers · 12 posts · Server hachyderm.ioOn a machine with 64 CPUs reading with hidefix is between 10x and 16x faster than native HDF5!!
And that is _including_ generating the index.
Going from 20 seconds to 1.8 seconds is pretty significant when reading a bunch of variables! 7s to 380ms, is the difference between waiting and immediately in processing!
This is for a file on a network disk.
The ultimate goal: Make this into a #xarray backend!
#xarray #rust #hidefix #hdf5 #data #met
gauteh · @gauteh
10 followers · 12 posts · Server hachyderm.iogauteh · @gauteh
9 followers · 6 posts · Server hachyderm.ioThe adventure continues in speedy HDF5 reading: reading directly into a target slice requires sorting by chunk so it is only read once. However, it turns out that grouping the segments within each chunk in a Vec of Vecs becomes very slow to deallocate! 2-3 seconds on a big variable. Better with a flat vector: https://github.com/gauteh/hidefix/pull/14/commits/f1cb9313c0828c668d95c2edd4b648d352f5408d?diff=unified&w=0#diff-c81afe1c3a0f0a1fea2d97a4071ac87058b7fd2fe843cb5855f181d7633e979fR501 #rust #optimize #hdf5 #data
Mark Kittisopikul · @markkitti
229 followers · 98 posts · Server fosstodon.orgI'm preparing for a #hdf5 sprint to meet 1.14 merge window for #H5Dchunk_iter consistency. This function allows you to efficiently iterate through chunk information such as the chunk address and compression filter state.
PR approved. Backporting to 1.12 required.
https://github.com/HDFGroup/hdf5/pull/2074
I might even try backporting to 1.10.
https://forum.hdfgroup.org/t/backporting-h5dchunk-iter-to-1-12-and-1-10/9971
openPMD · @openPMD
0 followers · 1 posts · Server mast.hpc.socialHey there 👋
Did someone say #standardization, #FAIRdata and #HPC?
Check our our community work on productive data #compatibility, leveraging leading HPC file formats such as #ADIOS and #HDF5!
#standardization #fairdata #hpc #compatibility #adios #hdf5
Kostas Andreadis · @kandreadis
5 followers · 12 posts · Server fediscience.orgTrying to reproject some #nasa #smap global soil moisture data, using #gdal the one solution that seems to consistently work requires setting the extent when extracting from the #hdf5 file https://gis.stackexchange.com/a/253959
Fred · @fred
85 followers · 57 posts · Server m.baldhead.beI still miss some #NetCDF #zarr #GeoTIFF #COG #geoparquet #HDF5 #OGC #geospatial people.
Anyone out there? :)
#netcdf #zarr #geotiff #cog #geoparquet #hdf5 #ogc #geospatial
Yann :python: · @nobodyinperson
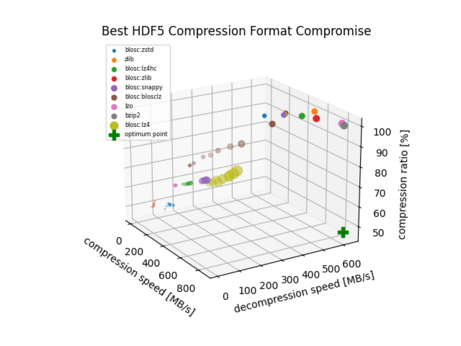
86 followers · 316 posts · Server fosstodon.org@thfriedrich I benchmarked the different compression algorithms in #HDF5 once if you're interested: https://gitlab.com/-/snippets/2043808
With the metric I use there (distance to optimum 'fast and small'), blosc:lz4 is the best compromise.
I still hit a wall with #HDF5 at some point though, I guess the compression prevented something from being done, I don't remember...
Also, #NetCDF4 is a subtype of #HDF5 so you'll feel familiar.
Thomas Friedrich · @thfriedrich
47 followers · 84 posts · Server fosstodon.org@nobodyinperson thanks for the insight. 🙂 I’ll have a look at #MQTT and #NetCDF4 which are both new to me. Working in science, so far I used mostly #hdf5 for larger data, which I think has a + for #openData & #openScience since there’s easy interfaces for most programming languages. Compression is decent I believe.
#mqtt #NetCDF4 #hdf5 #opendata #openscience
Arne Babenhauserheide · @ArneBab
397 followers · 9174 posts · Server rollenspiel.social#HDF5 and #Feather performance: storing and reading columnar float
https://github.com/fizban99/hdf_vs_feather/blob/main/hdf_vs_feather.ipynb
#benchmark
Feather writes faster, HDF5 reads faster at large sizes.