Prathyusha Konda · @prats
210 followers · 352 posts · Server fediscience.orgRT @LabListon@twitter.com
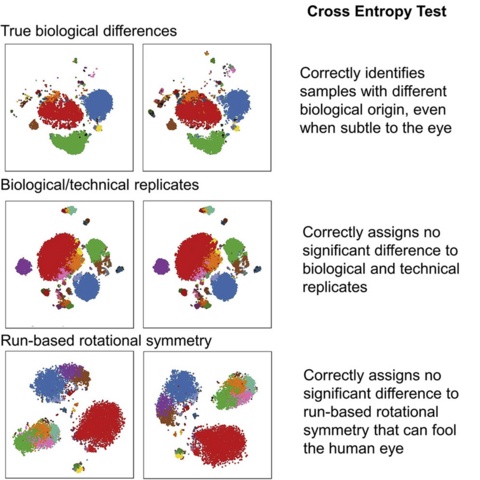
We have an exciting #computational paper out in @CellRepMethods@twitter.com. Ever use a #tSNE or #UMAP in #scSeq, #flowcytometry or #masscytometry? It doesn't have to be just a pretty picture anymore - we've developed a statistical test to check for differences. 1/6
https://www.cell.com/cell-reports-methods/fulltext/S2667-2375(22)00295-8
🐦🔗: https://twitter.com/LabListon/status/1615673972850413569
#masscytometry #flowcytometry #scseq #umap #tsne #computational
Dom Somma · @dom_somma
22 followers · 37 posts · Server genomic.socialRT @LabListon@twitter.com
We have an exciting #computational paper out in @CellRepMethods@twitter.com. Ever use a #tSNE or #UMAP in #scSeq, #flowcytometry or #masscytometry? It doesn't have to be just a pretty picture anymore - we've developed a statistical test to check for differences. 1/6
https://www.cell.com/cell-reports-methods/fulltext/S2667-2375(22)00295-8
🐦🔗: https://twitter.com/LabListon/status/1615673972850413569
#computational #tsne #umap #scseq #flowcytometry #masscytometry
C. Harris Floudas · @chfloudas
70 followers · 339 posts · Server mstdn.socialRT @LabListon@twitter.com: We have an exciting #computational paper out in @CellRepMethods. Ever use a #tSNE or #UMAP in #scSeq, #flowcytometry or #masscytometry? It doesn't have to be just a pretty picture anymore - we've developed a statistical test to check for differences. 1/6
https://www.cell.com/cell-reports-methods/fulltext/S2667-2375(22)00295-8 https://twitter.com/LabListon/status/1615673972850413569/photo/1
#masscytometry #flowcytometry #scseq #umap #tsne #computational
Adrian Liston · @ListonLab
133 followers · 138 posts · Server mastodon.auIf you can already use R, you can skip the tutorial and get the repository straight from GitHub. We have separate scripts optimised for #flowcytometry / #masscytometry or #scSeq. As well as the stats there are nice data visualisation tools included. 4/6 https://github.com/AdrianListon/Cross-Entropy-test
#flowcytometry #masscytometry #scseq
Adrian Liston · @ListonLab
133 followers · 135 posts · Server mastodon.auWe have an exciting #computational paper out in @CellRepMethods@twitter.com. Ever use a #tSNE or #UMAP in #scSeq, #flowcytometry or #masscytometry? It doesn't have to be just a pretty picture anymore - we've developed a statistical test to check for differences. 1/6
https://www.cell.com/cell-reports-methods/fulltext/S2667-2375(22)00295-8
#computational #tsne #umap #scseq #flowcytometry #masscytometry
Rohit Farmer, Ph.D. · @swatantra
236 followers · 162 posts · Server fosstodon.orgThis paper describes HDStIM. HDStIM is a method for identifying responses to experimental #stimulation in #massCytometry or #flowCytometry that uses a #highDimensional analysis of measured parameters and can be performed with an end-to-end #unsupervised approach.
#stimulation #masscytometry #flowcytometry #highDimensional #unsupervised