Stephen Matheson🌵🌲 · @sfmatheson
709 followers · 696 posts · Server fediscience.org#Multiplex
#Imaging
#MachineLearning
calling this Combplex -- combinatorial multiplexing
validated, then adapted to mass-based approaches (MIBI-TOF)
#MassSpec
How does the method work? It learns the compression matrix
2/2
#massspec #machinelearning #imaging #multiplex
PastelBio · @pastelbio
175 followers · 1927 posts · Server mstdn.scienceRT @Matrix_Science: You don't need a fancy new search engine to identify peptides from chimeric spectra. Just process the raw file in Mascot Distiller and search with Mascot Server. #proteomics #MassSpec #MassSpectrometry #Bioinformatics
https://www.matrixscience.com/blog/identifying-peptides-from-chimeric-spectra-dda-dia.html
#proteomics #massspec #massspectrometry #bioinformatics
PastelBio · @pastelbio
169 followers · 1756 posts · Server mstdn.scienceRT @: Real-Time Spectral Library Matching for Sample Multiplexed Quantitative Proteomics #JProteomeRes #MassSpec
Johannes Rainer · @jorainer
260 followers · 254 posts · Server fosstodon.orgAdded support for #MassSpec spectra comparisons using the spectral entropy similarity score to our #Spectra @bioconductor package (dev version):
Thanks Yuanyue Li for the #msentropy #rstats :rstats: package and support! 🥳
#massspec #spectra #msentropy #rstats
AutoVectis · @autovectis
35 followers · 35 posts · Server mastodon.org.ukWe have a free (already paid for) space available at this year's Ardgour Symposium! More details here - www.emsg-ardgour.site If you can get yourself to Aberdeen on the 20th August, then you could be enjoying a week of mass spec and related discussions, in the Highlands and with a free bar! #teammassspec #massspec #lipidomics #metabolomics #proteomics #ionization
#teammassspec #massspec #lipidomics #metabolomics #proteomics #ionization
PastelBio · @pastelbio
164 followers · 1673 posts · Server mstdn.scienceRT @: What is the most popular / robust/ reliable tool out there for analyzing cross-linking #massspec XL-MS data ?
Asking for a friend.
#massspec #proteomics #tools #resources
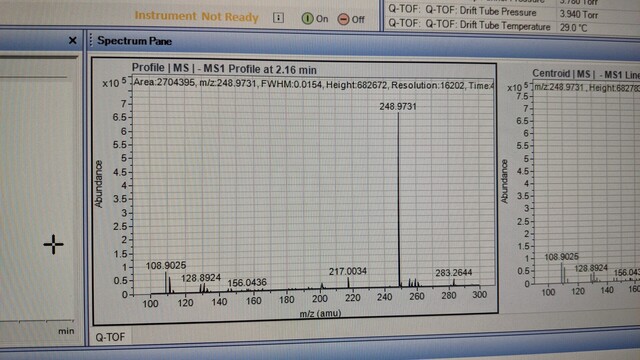
Sarah Hancock · @sarahehancock
148 followers · 3 posts · Server mstdn.scienceJust in case anyone in #TeamMassSpec is curious, the likely culprit of my 248.9 m/z ion from this thread: https://mstdn.science/@sarahehancock/110818463525422168 is free cyanide reacting with the gold seals of the HPLC pump. I will have to try a few different brands of solvent to see if I can ameliorate this issue. More info also available here: https://community.agilent.com/technical/lcms/f/forum/5480/how-do-we-get-rid-of-identify-contamination-with-m-z-248-9737-negative-mode.
#TeamMassSpec #metabolomics #lipidomics #massspectrometry #massspec
Sarah Hancock · @sarahehancock
147 followers · 2 posts · Server mstdn.scienceHey #TeamMassSpec, I'm having issues with a contaminant ion @ 248.9 m/z in -ve ion mode when running 90% ACN 10 mM AmOAc at pH 9. Found a thread on research gate suggesting it comes from the ACN but have tried two brands now with no joy. Has anyone else had this problem?
Research gate link: https://researchgate.net/post/What-is-this-contaminant-with-m-z-2489718
ACN brands tried: Fisher Sci Optima, Supelco LiChroSolv. LiChroSolv has a lot less of it than Fisher Sci.
#TeamMassSpec #metabolomics #lipidomics #massspectrometry #massspec
PastelBio · @pastelbio
164 followers · 1618 posts · Server mstdn.scienceRT @: Always good to see new #massspec tools available.Dear #proteomicscommunity and #TeamMassSpec, I am happy to share HowDirty, an R package that facilitates evaluating and reporting molecular contaminants in LC-MS experiments.
https://authorea.com/users/643346/articles/656759-howdirty-an-r-package-to-evaluate-molecular-contaminants-in-lc-ms-experiments…
#massspec #proteomicscommunity #TeamMassSpec
PastelBio · @pastelbio
158 followers · 1504 posts · Server mstdn.scienceRT @: Anyone have advice on how they normalize TMT channel data when the samples are from a pulldown? Normalization methods assume that peptide abundances should be broadly equal across samples...
#TeamMassSpec #massspec #proteomics
wfondrie · @wfondrie
200 followers · 66 posts · Server genomic.socialI gave a talk on Depthcharge, our #deeplearning toolkit for #massspec data at the Cascadia #Proteomics Symposium this week. Here’s my slides in case you missed it, or want to see them again!
#deeplearning #massspec #proteomics
jobRxiv · @jobRxiv
936 followers · 2880 posts · Server mas.toPostdoc, Kidney metabolism and metabolomics
Aarhus University
See the full job description on jobRxiv: https://jobrxiv.org/job/aarhus-university-27778-postdoc-kidney-metabolism-and-metabolomics/?feed_id=49965
#ScienceJobs #hiring #research #massspec
Aarhus University #Denmark #PhDStudent #PostdoctoralFellow
https://jobrxiv.org/job/aarhus-university-27778-postdoc-kidney-metabolism-and-metabolomics/?feed_id=49965
#postdoctoralfellow #PhDstudent #denmark #massspec #research #hiring #ScienceJobs
jobRxiv · @jobRxiv
934 followers · 2839 posts · Server mas.toPostdoc, Kidney metabolism and metabolomics
Aarhus University
See the full job description on jobRxiv: https://jobrxiv.org/job/aarhus-university-27778-postdoc-kidney-metabolism-and-metabolomics/?feed_id=49678
#ScienceJobs #hiring #research #massspec
Aarhus University #Denmark #PhDStudent #PostdoctoralFellow
https://jobrxiv.org/job/aarhus-university-27778-postdoc-kidney-metabolism-and-metabolomics/?feed_id=49678
#postdoctoralfellow #PhDstudent #denmark #massspec #research #hiring #ScienceJobs
PastelBio · @pastelbio
153 followers · 1344 posts · Server mstdn.scienceRT @: JULY 15 << The Abstract Deadline is fast approaching for the 2ND INTERNATIONAL TOP-DOWN PROTEOMICS SYMPOSIUM. Submit your work now at> https://tdp2023.topdownproteomics.org/submit-an-abstract/… #proteomics #ASMS #genomics #massspec #proteoform
#proteomics #ASMS #genomics #massspec #proteoform
PastelBio · @pastelbio
147 followers · 1295 posts · Server mstdn.scienceRT @: Hey #TeamMassSpec - question re: low sample stage-tip fractionation. Anyone have any experience with it? We have some POROS 20 R2 packing. How much (~mg?) do you load into a 200µL tip?
#TeamMassSpec #massspec #proteomics
PastelBio · @pastelbio
136 followers · 1169 posts · Server mstdn.scienceRT @: 20,001 DOWNLOADS - The Human Proteoform Project seems to be of interest to quite a few people... #proteomics #proteoforms #genomics #massspec
https://science.org/doi/10.1126/sciadv.abk0734…
#proteomics #proteoforms #genomics #massspec
Dr Micha Campbell · @michcampbell
455 followers · 925 posts · Server fediscience.orgCan someone who knows something about mass spectrometry explain why the Agilent 8900 is called a triple quadrupole, when I'm fairly sure there are only two quads??
#massspec #massspectrometry #labwork
PastelBio · @pastelbio
132 followers · 1125 posts · Server mstdn.scienceRT @: JOB OPPORTUNITY!
Looking for a post-doc developing multiplexed targeted #massspec assays to support clinical translation? Look no further!
#massspec #TeamMassSpec #proteomics
PastelBio · @pastelbio
131 followers · 1041 posts · Server mstdn.scienceMT @: A great opportunity for #SingleCell omics, #MassSpec, & #imaging #MassSpec researchers to publish their advances on pushing tech towards single-cell analyses, sensitivity & resolution JPR is not only about proteins -- metabolites, lipids, glycans, drugs, exposome, all welcome!Theo @thalexandrov and I will serve as editors for a Special Issue of @JProteomeRes Single-Cell Omics
Submissions before January 31, 2024 can be included in the special issue.
#singlecell #massspec #imaging
Johannes Rainer · @jorainer
246 followers · 242 posts · Server fosstodon.orgLecture on proteomics by @lgatto at #csama 2023 including an overview on the :rstats: package ecosystem from #RforMassSpectrometry #MassSpec
#csama #rformassspectrometry #massspec