Fatih Demir · @kabalak
53 followers · 98 posts · Server mstdn.scienceAnybody working with MaxDIA on semispecific searches? Importing a FragPipe-generated library.tsv does not seem to work beyond the "Peptide FDR1" step. The discovery libraries publicly available only feature specific, trypsin digestions and nothing semi-specific. #MaxQuant #MaxDIA (things you ought to not do at all... semi-specific with MaxDIA).
Brian Hampton · @ionurchin
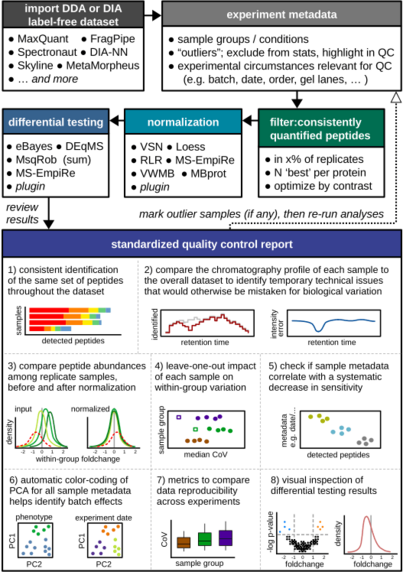
25 followers · 47 posts · Server mastodon.socialSomething new to test out for the new year #MSDAP was just published. "The #MassSpectrometry Downstream Analysis Pipeline (MS-DAP) is an open-source R package for analyzing and interpreting #label-free #proteomics datasets". It takes output from many upstream search pipelines like #MSFragger, #MetaMorpheous, #DIA-NN, #Spectronaut, #MaxQuant etc. and then applies a range of QC and statistical workflows.
https://pubs.acs.org/doi/10.1021/acs.jproteome.2c00513
#msdap #massspectrometry #label #proteomics #msfragger #metamorpheous #dia #spectronaut #maxquant