Kurianlab · @kurianlab
80 followers · 509 posts · Server sciencemastodon.comRT @vizgen_inc
🇫🇷We're excited to sponor Applications of Single Cell Genomics Across Development & Immune Diseases at @NantesUniv! Join us April 4th at 15:30 GMT+1 to discover #MERSCOPE & #MERFISH for high-quality #SpatialTranscriptomics.
Check out more Vizgen events: https://hubs.ly/Q01J6K2S0
#merscope #merfish #spatialtranscriptomics
Kurianlab · @kurianlab
78 followers · 443 posts · Server sciencemastodon.comRT @vizgen_inc
🧠Looking for some brainy fun this week? Check out the original #MERSCOPE #MERFISH Mouse Brain Receptor Map!🐭
-483 gene panel
-734,969 total cells imaged
-554,802,908 total transcripts detected
-GPCRs & RTKs
Explore today: https://hubs.ly/Q01GFS8T0🎣
#merscope #merfish #brainweek #neurotwitter
Kurianlab · @kurianlab
78 followers · 427 posts · Server sciencemastodon.comRT @vizgen_inc
📣We are excited to share a preprint from our brilliant scientific co-founder Dr. Xiaowei Zhuang's lab at @Harvard!
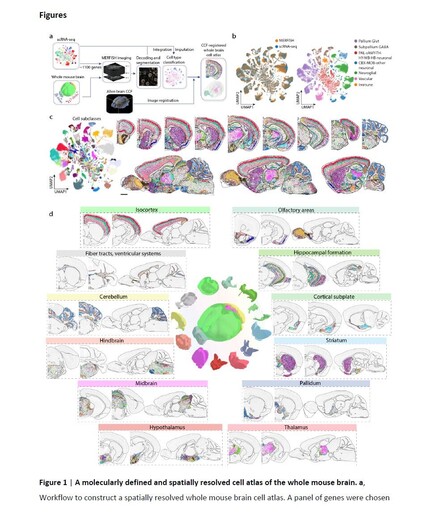
Meng Zhang's amazing team used #MERFISH to create a #CellAtlas of the whole mouse brain- https://hubs.ly/Q01FWLM10
🐭🧠👩🔬
#merfish #cellatlas #iwd2023 #embraceequity #WomenInScience
Kurianlab · @kurianlab
77 followers · 393 posts · Server sciencemastodon.comRT @vizgen_inc
#SpatialMultiomic measurements that combine transcript + protein detection + location are critical in understanding complex #biology.
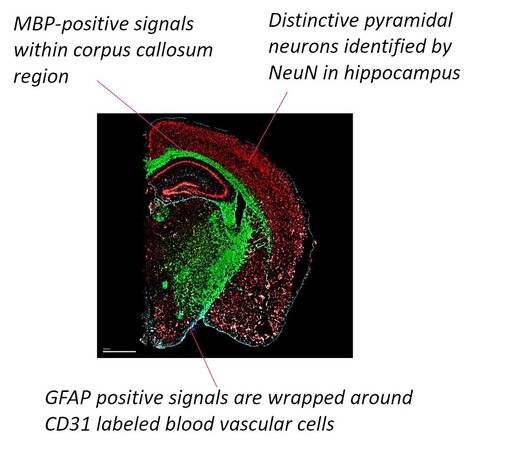
This #MERSCOPE masterpiece shows the co-detection of 5 proteins + 468 #MERFISH gene panel on mouse brain.
Happy National Protein Day!
🐭🧠💪
#spatialmultiomic #biology #merscope #merfish
Hao Yin · @HaoYin
222 followers · 255 posts · Server qoto.orgCOMMOT, COMMunication analysis by Optimal Transport, a versatile tool infer cell–cell communication in #SpatialTranscriptomics 🤠
Compatible with #MERFISH #Visium seqFISH+ #STARmap
vs #CellChat #Giotto #CellPhoneDBv3
Dr. Qing Nie lab Nature Methods 2023
https://www.nature.com/articles/s41592-022-01728-4
#CellChat #giotto #CellPhoneDBv3 #merfish #visium #Starmap #SpatialTranscriptomics
Ming 'Tommy' Tang · @tangming2005
19 followers · 3 posts · Server genomic.socialIf you work on the spatial transcriptome data. Our tool may help you https://monkeybread.readthedocs.io/en/latest/ #merfish #spatial
kevin lebrigand · @kevinlebrigand
8 followers · 4 posts · Server genomic.socialRT @vizgen_inc@twitter.com
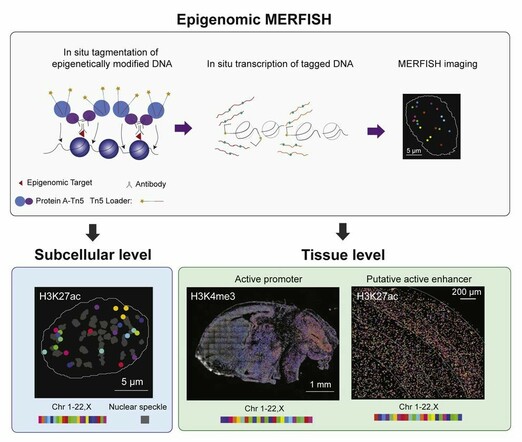
New #MERFISH publication alert! Dr. Xiaowei Zhuang's @Harvard@twitter.com research team developed #EpigenomicMERFISH! Check out the open access publication available from @CellCellPress@twitter.com: https://hubs.ly/Q01v3jSG0
#Fascinating #Epigenomics #MERFISH #Brilliant
🐦🔗: https://twitter.com/vizgen_inc/status/1600135473770496000
#merfish #epigenomicmerfish #fascinating #Epigenomics #brilliant
Haojia Wu · @haojiawu
24 followers · 8 posts · Server mastodon.haojia-wu.comJoint Sparse method for Imaging Transcriptomics (JSIT) for decoding imaging spatial transcriptomics data such as MERFISH. Better performance than the original pipeline (MERlin) according to the authors. #SpatialTranscriptomics #matlab #merfish #bioinformatics #tools
https://www.biorxiv.org/content/10.1101/2022.11.22.517523v1.full
#spatialtranscriptomics #matlab #merfish #bioinformatics #tools
Haojia Wu · @haojiawu
21 followers · 8 posts · Server mastodon.jiaworkspace.comJoint Sparse method for Imaging Transcriptomics (JSIT) for decoding imaging spatial transcriptomics such as MERFISH. Better performance than the original pipeline (MERlin) according to the authors. #SpatialTranscriptomics #matlab #merfish #bioinformatics #tools
https://www.biorxiv.org/content/10.1101/2022.11.22.517523v1.full
#spatialtranscriptomics #matlab #merfish #bioinformatics #tools
Anshul Kundaje · @akundaje
1078 followers · 260 posts · Server genomic.socialRT @vizgen_inc@twitter.com
#30DayMapChallenge Day 27- #Heatmap
#MERFISH mouse brain map. The interactive heatmap enables the visualization of gene expression across Leiden clusters and single-cells!
https://hubs.ly/H0_V5260
#observable_jupyter #deckgl #spatialtranscriptomics @googlecolab@twitter.com @observablehq@twitter.com
🐦🔗: https://twitter.com/vizgen_inc/status/1464626128484806663
#30daymapchallenge #heatmap #merfish #observable_jupyter #deckgl #spatialtranscriptomics