Thomas Haverkamp · @thomieh
9 followers · 14 posts · Server genomic.socialHi, currently looking at a #phage proposal. Does anybody know a #bioinformatics tool that can search for virus sequences in #metagenome assemblies?
#phage #bioinformatics #metagenome
NIOO-KNAW · @niooknaw
27 followers · 120 posts · Server social.edu.nlNew NIOO publication: Impact of the fungal #pathogen Fusarium oxysporum on the taxonomic and functional diversity of the common bean root #microbiome. #rhizosphere #endosphere #metagenome #metatranscriptome
https://doi.org/10.1186/s40793-023-00524-7
#pathogen #microbiome #rhizosphere #endosphere #metagenome #metatranscriptome
James Marsh · @marshomics
21 followers · 20 posts · Server fediscience.orgAnother windy, rainy morning in Germany but at least 30,000 of my metagenomes have been annotated since yesterday. Only 430,000 more to go.. ⏰ #microbiome #metagenome
naturepoker · @naturepoker
639 followers · 1005 posts · Server genomic.socialAccording to the new #metagenome paper, #viruses / #phages of non-aquatic environments only outnumber cells by about two-fold (major capsid protein and universal SCGs used as markers). So the new estimation of virus/phage count might be far closer to cells. Interesting. #microbiology
#microbiology #metagenome #viruses #Phages
PLOS Biology · @PLOSBiology
4864 followers · 1438 posts · Server fediscience.org#Viruses infecting #bacteria & #archaea are often identified via metagenomics, but information regarding #phage hosts is lost. @simroux_virus &co present iPHoP, a new tool to maximize host prediction for #metagenome-assembled virus genomes #PLOSBiology https://plos.io/41A5QvG
#plosbiology #metagenome #phage #archaea #bacteria #viruses
PLOS Biology · @PLOSBiology
4863 followers · 1426 posts · Server fediscience.org#Viruses infecting #bacteria & #archaea are often identified via metagenomics, but information regarding #phage hosts is lost. @simroux_virus &co present iPHoP, a new tool to maximize host prediction for #metagenome-assembled virus genomes #PLOSBiology https://plos.io/41A5QvG
#plosbiology #metagenome #phage #archaea #bacteria #viruses
James Fellows Yates · @jfy133
278 followers · 259 posts · Server genomic.social🚨 New pipeline release! 🚨
🦠 The nf-core/mag v2.3.0 (Red Cow) pipeline for #metagenome assembly and binning now includes CheckM and GUNC for improved #MAG completeness esimates, CONCOCT as an additional binner, and improved CAT reporting, alongside a range of bug fixes and improvements!
Thanks as always to all the contributors!
Victor · @Victor
40 followers · 377 posts · Server spore.social"...high-resolution spatio-temporal #sampling of the spring bloom in a #freshwater reservoir and describe a multitude of previously unknown taxa using #metagenome-assembled genomes of #eukaryotes, #prokaryotes, and #viruses in combination with a broad array of methodologies." https://pubmed.ncbi.nlm.nih.gov/36698172/
#sampling #freshwater #metagenome #eukaryotes #prokaryotes #viruses
Victor · @WWBugs
52 followers · 452 posts · Server mstdn.social"...high-resolution spatio-temporal #sampling of the spring bloom in a #freshwater reservoir and describe a multitude of previously unknown taxa using #metagenome-assembled genomes of #eukaryotes, #prokaryotes, and #viruses in combination with a broad array of methodologies." https://pubmed.ncbi.nlm.nih.gov/36698172/
#viruses #Prokaryotes #eukaryotes #metagenome #freshwater #sampling
Francis Ouellette · @bffo
79 followers · 12 posts · Server mastodon.socialFrom @plos #Computational #biology | Ten simple rules for investigating (meta)genomic data from environmental ecosystems | #TSRPLOSCB #metagenome #editorial #bioinformatics | I like their Fig. 1, and I like all of the rules, but I am partial to Rule 7: Databases are the key to everything | https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1010675
#computational #biology #tsrploscb #metagenome #editorial #bioinformatics
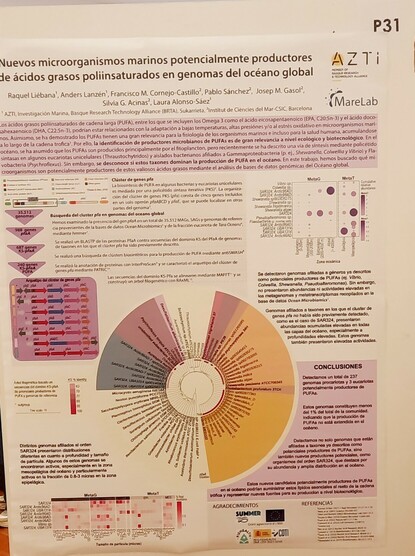
Anders Lanzén · @lanzen
133 followers · 116 posts · Server fediscience.org@raliegar unfortunately could not make it to the #MikrobioGune meeting here in Bilbao, but I'm proud to stand in and present her poster below about our daring hunt for #omega3 and other #PUFA producing microbes in the #mesopelagic and other marine strata, using public and in-house #metagenome s.
#metagenome #mesopelagic #pufa #omega3 #mikrobiogune
Vivek Mutalik · @vivek_mutalik
455 followers · 310 posts · Server fediscience.orgDeep divergence and genomic diversification of gut #symbionts of neotropical stingless bees | bioRxiv
#metagenome #microbiome #bees #symbionts
Rob Edwards · @linsalrob
168 followers · 30 posts · Server genomic.socialFantastic talk from @wwood@twitter.com about their Sandpiper tools for #metagenome analysis https://sandpiper.qut.edu.au/ and how they have been using it for strain level metagenomics