NFDI4Microbiota · @NFDI4Microbiota
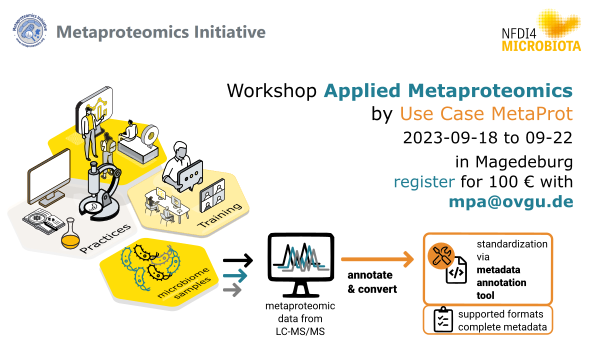
47 followers · 20 posts · Server nfdi.socialThere are only a few seats left for this #metaproteomics hands-on training workshop run by the NFDI4Microbiota Use Case MetaProt!
🗓️18th to 22nd September @OVGUpresse #Magdeburg.
Registration fee: 100 € (incl. coffee and snacks)
Further details: https://t1p.de/6ihj2
PastelBio · @pastelbio
166 followers · 1724 posts · Server mstdn.scienceRT @: The @MyABRF IPRG study on #Metaproteomics (iPRG-2020) is now published in Journal of Biomolecular Techniques. https://jbt.pubpub.org/pub/01nicjgy/release/1…
PastelBio · @pastelbio
165 followers · 1716 posts · Server mstdn.scienceRT @: Metaproteomics study from 2020 a great read for everyone in the field. The @MyABRF IPRG study on #Metaproteomics (iPRG-2020) is now published in Journal of Biomolecular Techniques. https://jbt.pubpub.org/pub/01nicjgy/release/1…
IMPACTT Microbiome · @IMPACTT
83 followers · 45 posts · Server mstdn.scienceIMPACTT Mentee Speaker Profile for #HavingIMPACTT2023: Meet Dr. Caitlin Simopoulos, Computational Biologist at Roche Canada who will speak about highlighting the power of #metaproteomics through CAMPI3, a #microbiome functional benchmarking study
#microbiota #conference #symposium #science #benchmark #study #computational #biology
#havingimpactt2023 #metaproteomics #microbiome #microbiota #conference #symposium #Science #benchmark #study #computational #biology
PastelBio · @pastelbio
130 followers · 993 posts · Server mstdn.scienceRT @: Join LS2 Proteomics Webinar Mini Series on the 22 June 2023 (15:00 Swiss time) together with Jana Seifert - University of Hohenheim and Tim Van Den Bossche - Ghent University, founding members of the Metaproteomics Initiative!
https://meetings.ls2.ch/webinar-series
#metaproteomics #proteomics
PastelBio · @pastelbio
119 followers · 466 posts · Server mstdn.scienceRT @kalonji_08: Hey #proteomics community. I’m looking for some #massspec tutorials especially with regards to #metaproteomics and @proteogenomics analysis. Any assistance would be appreciated Thanks #Bioinformatics
#proteomics #massspec #metaproteomics #bioinformatics
Mol Syst Biol · @MolSystBiol
217 followers · 27 posts · Server sciencemastodon.comYeast + lactic acid bacteria co-cultures grow on amino acid-poor lactose medium, where none of these species can grow alone. 13C labelling & #metaproteomics show amino acid & carbon flow between community members ➡️ http://bit.ly/3Ik9yCj
from Kiran Patil, Cambridge University
#microbialcommunity #SystemsBiology
#metaproteomics #microbialcommunity #systemsbiology
Akos T Kovacs · @EvolvedBiofilm
432 followers · 1600 posts · Server mstdn.scienceRT @Manuel_Kleiner
Wow, almost two years later our article on Protein-SIP is finally published https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-022-01454-1
Thank you everyone for you hard work on this one and also thank you to the reviewers who really helped to improve the article #metaproteomics #NCStateMicrobiome @marc_strous https://twitter.com/Manuel_Kleiner/status/1377281990907588608
#metaproteomics #ncstatemicrobiome
Vijini Mallawaarachchi · @vijinim
22 followers · 6 posts · Server genomic.socialOur first publication of 2023 is now out in Frontiers in Microbiology. We study the composition and metabolism of microbial communities in Daqu, the Chinese liquor fermentation starter.
#Bioinformatics #metaproteomics #research #analysis
https://www.frontiersin.org/articles/10.3389/fmicb.2022.1098268/full
#bioinformatics #metaproteomics #research #analysis
Vijini Mallawaarachchi · @vijinim
22 followers · 6 posts · Server genomic.socialThe first publication of 2023 is now out in Frontiers in Microbiology. We study the composition and metabolism of microbial communities in Daqu, the Chinese liquor fermentation starter.
#Bioinformatics #metaproteomics #research #analysis
https://www.frontiersin.org/articles/10.3389/fmicb.2022.1098268/full
#bioinformatics #metaproteomics #research #analysis
Akos T Kovacs · @EvolvedBiofilm
175 followers · 281 posts · Server mstdn.scienceRT @ScienceCluster
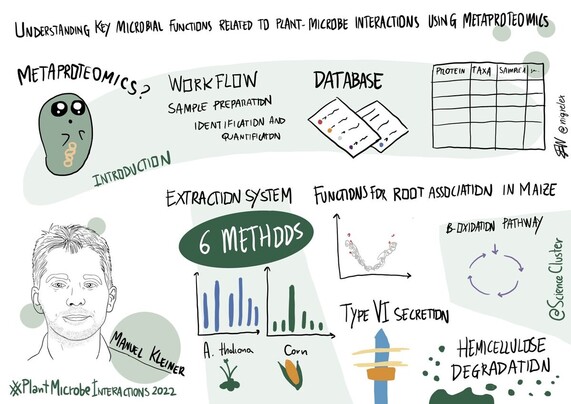
Continuing the session on methods and #technologies for #plant #microbiome interrogation with @Manuel_Kleiner 🌱
The talk was on understanding key microbial functions related to #plant-microbe interactions #metaproteomics 🪴
#PlantMicrobeInteractions2022
@novonordiskfond
#technologies #plant #microbiome #metaproteomics #PlantMicrobeInteractions2022
Akos T Kovacs · @EvolvedBiofilm
168 followers · 260 posts · Server mstdn.scienceManuel Kleiner @Manuel_Kleiner (from @NCState @NCStateCALS; #NNF_INTERACT #NNF_InRoot) on understanding key microbial functions related to plant-microbe interactions using metaproteomics
#NNF_INTERACT #NNF_InRoot #metaproteomics #PlantMicrobeInteractions2022