Una · @sciolato
74 followers · 844 posts · Server mstdn.scienceNew to me #nanopore stuff, sure other folks know this already. If I chop off the first 100 bp from reads, the average quality goes way up. Filtering greater than q20 gave me 20k more reads if I did the chop.
Christos Argyropoulos MD, PhD · @ChristosArgyrop
371 followers · 575 posts · Server mstdn.scienceLooking forward to chat at #KidneyWeek @ASNKidney , where we will be showing our pilot work (mostly done by @MorganMacKenz12@twitter.com, I just abused the Xeons for the #bioinformatics) regarding the role of @nanopore #nanopore sequencers in the discovery of kidney disease biomarkers.
#kidneyweek #bioinformatics #nanopore
Jennifer Malsom · @jmalsom
74 followers · 20 posts · Server genomic.socialRegister today for our Nanopore Day in Copenhagen to hear the latest technical updates from Oxford Nanopore Technologies as well as talks from local and international scientists about their work using nanopore technology in human translational and clinical research.
#genomics #clinicalgenomics #nanopore #sequencing
tchrisboles · @tchrisboles
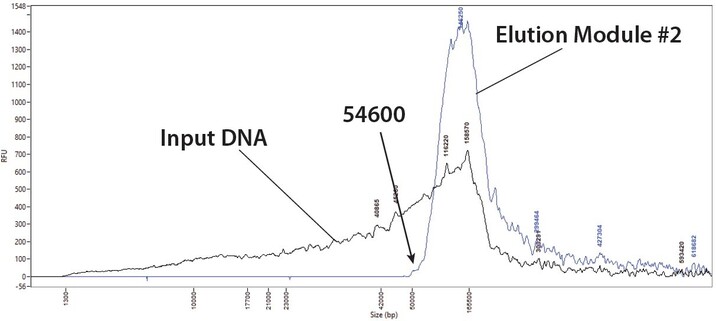
46 followers · 117 posts · Server genomic.socialHey #nanopore users, check out our new application note on using SageHLS for size selection at higher MW cutoff values: https://sagescience.com/application-notes/
Chris_Faulk_bio · @Chris_Faulk_bio
16 followers · 29 posts · Server genomic.socialTeresita Porter 🙋🏻♀️ · @DNAdataPhile
121 followers · 489 posts · Server ecoevo.social"Rapid species discovery and identification with real-time barcoding facilitated by ONTbarcoder 2.0 and Oxford Nanopore R10.4"
https://www.biorxiv.org/content/10.1101/2023.06.26.546538v2
The accompanying video is cool
https://twitter.com/RudolfMeier15/status/1674801657287413760?s=20
Samuel Nestor Meckoni · @samnm
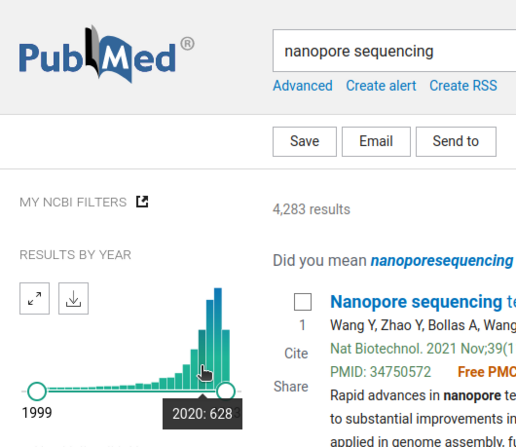
2 followers · 2 posts · Server mstdn.scienceFunny detail found in PubMed searching for "nanopore sequencing": The total number of search results from 2020 equals the ones from 2023 as of today.
The nanopore sequencing technology is currently rising. When will it decline? When every genome of every known species is sequenced, or when another technology outpaces it?
#nanopore #sequencing #pubmed
Samuel Nestor Meckoni · @samnm
2 followers · 2 posts · Server mstdn.scienceFunny detail found in PubMed searching for "nanopore sequencing": The total number of search results from 2020 equals the ones from 2023 as of today. The Nanopore sequencing technology is currently rising. When will it decline? When every genome of every known species is sequenced, or when another technology outpaces it?
#nanopore #sequencing #pubmed
Christos Argyropoulos MD, PhD · @ChristosArgyrop
322 followers · 420 posts · Server mstdn.science@mdperry Hope to integrate those in our pipeline for #nanopore https://www.biorxiv.org/content/10.1101/2022.12.16.520507v1. Just finished rewriting from #Python to #perl and looking for something other than blast (I was unpleasantly surprised by the fragility of kmer seeding methods in the presence of sequencing errors).
Phys.org · @physorg_bot
502 followers · 14667 posts · Server social.platypush.techReferenced link: https://phys.org/news/2023-06-extreme-dna-resolution-spatially-multiplexed.html
Discuss on https://discu.eu/q/https://phys.org/news/2023-06-extreme-dna-resolution-spatially-multiplexed.html
Originally posted by Phys.org / @physorg_com: http://nitter.platypush.tech/physorg_com/status/1670808664788156417#m
Extreme DNA resolution: Spatially multiplexed single-molecule translocations through a #nanopore at controlled speeds @epfl_en @NatureNano https://phys.org/news/2023-06-extreme-dna-resolution-spatially-multiplexed.html
Stephen P. Cohen, Ph.D. · @spcohen
28 followers · 19 posts · Server mstdn.scienceAnyone have any life changing tips for a first-time #nanopore user? I will be doing some MinION sequencing over the next few weeks.
Christos Argyropoulos MD, PhD · @ChristosArgyrop
317 followers · 412 posts · Server mstdn.scienceAnd this simulation #rstats code, shows why one must use use very short seeds when using #blast to map #nanopore #transcriptomic #RNAseq data (Q10 quality scores are not unheard of, in such datasets).
Take home message, one has got to work for their database searches for long read platforms
(R code in the alt description)
#rstats #blast #nanopore #transcriptomic #rnaseq
Human Technopole · @humantechnopole
312 followers · 87 posts · Server mstdn.science🔬 What are the opportunities and the challenges of nanopore sequencing technology? We talked about it with Philipp Rescheneder @nanopore@genomic.social @nanopore@bird.makeup
“One of the things that I like about nanopore sequencing is that it’s very flexible”
#sciencemastodon #science #lifesciences #nanopore #humantechnopole #sequencing
https://www.youtube.com/watch?v=6awqLEHshrY&list=PLZAXXKHOAZejmAUgTARUfsXbREH0qsd9G&index=11
#MeetTheSpeaker #sciencemastodon #Science #lifesciences #nanopore #humantechnopole #sequencing
Human Technopole · @humantechnopole
310 followers · 84 posts · Server mstdn.science🔬 Hazuki Takahashi from RIKEN IMS explains how nanopore long-read #sequencing technology can change research on the #humangenome
#sciencemastodon #science #lifesciences #humantechnopole #nanopore @nanopore@genomic.social @nanopore@bird.makeup @riken_en
#MeetTheSpeaker #sequencing #humangenome #sciencemastodon #Science #lifesciences #humantechnopole #nanopore
Joseph Guhlin · @josephguhlin
524 followers · 89 posts · Server sci.kiwi#Minimap2 #Rust library release: minimap2 v0.1.13+minimap2.2.26
* Latest minimap2 stable release (2.26)
* with_seq function
* hopefully compiles for MacOS / aarch64.
* Still struggling with SSE, I recommend using simde feature
*Soft-clipping properly in CIGAR strings but use caution. Happy to work on it more.
https://github.com/jguhlin/minimap2-rs
https://crates.io/crates/minimap2
And the -sys crate
https://crates.io/crates/minimap2-sys
#minimap2 #rust #bioinformatics #genomics #nanopore #PacBio
Joseph Guhlin · @josephguhlin
518 followers · 55 posts · Server sci.kiwiHopefully not taking the server down due to a lack of inodes.... #bioinformatics #genomics #nanopore
#nanopore #genomics #bioinformatics
Christos Argyropoulos MD, PhD · @ChristosArgyrop
312 followers · 374 posts · Server mstdn.scienceAlright, #bioinformatics folk, which multiple sequence #alignment and #clustering packages should I add to my #transcriptomic @nanopore workflow?
Linclust, uclust/ cd-hit are candidates for clustering.
Candidates for MSA: clustal/kalign/hmmalign.
Other suggestions? #nanopore #sequencing
#bioinformatics #alignment #clustering #transcriptomic #nanopore #sequencing
Martin Smith · @Martinalexsmith
404 followers · 316 posts · Server genomic.socialA substantial base calling quality improvement observed beta testing the #nanopore RNA004 direct #rnaseq protocol.
This is empirical data from commercial, multi-tissue RNA after poly(A) selection, aligned to hg38 with miminap2 -a -x splice --secondary=no.
Alignments (especially at splice junctions) look exceptional; I could see potential RNA modifications directly from basecalling output as mutations in IGV
Robert Castelo · @robertclab
98 followers · 371 posts · Server genomic.socialRT @hagentilgner
Andrey & Alla put out the newest version of Isoquant
(https://www.nature.com/articles/s41587-022-01565-y); Results unchanged but memory usage down ~10-15 fold and disk usage down ~5-fold;
https://github.com/ablab/IsoQuant/releases/tag/v3.2.0
#LongRead #RNA @iRnaCosi
#nanopore #pacbio #genomics
#Transcripomics #RNAseq #Splicing
#longread #RNA #nanopore #PacBio #genomics #transcripomics #rnaseq #splicing
Thomas Sandmann · @thomas_sandmann
287 followers · 583 posts · Server genomic.socialVery cool preprint by Adam P Cribbs's group: "Correcting PCR amplification errors in unique molecular identifiers to generate absolute numbers of sequencing molecules." https://www.biorxiv.org/content/10.1101/2023.04.06.535911v1 with lots of innovative ideas to a) measure and b) correct PCR induced errors in RNA-seq experiments. #rnaseq #ngs #singlecells #compbio #illumina #nanopore
#rnaseq #ngs #singlecells #compBio #illumina #nanopore