Bioconductor · @bioconductor
369 followers · 48 posts · Server genomic.social🎓 #biocsmorgasbord! starts today! New workshop added ✨ Learn "Introduction to SpatialOmicsOverlay" with the #NanoString #GeoMx Team 🧬💻
📅 May 22-26
➡️ https://gallantries.github.io/video-library/modules/bioconductor
#Bioinformatics #RStats #SpatialOmics
#biocsmorgasbord #nanostring #geomx #bioinformatics #RStats #spatialomics
David Mason · @dn_mason
171 followers · 151 posts · Server mas.toYou might also find me hanging out at the #StandardBioTools, #NanoString or #Akoya booths.
I also contributed to a cool poster on RNA / Protein #multiplex in Head & Neck Cancer if that's your jam.
#ish #rnascope #aacr23 #Multiplex #akoya #nanostring #standardbiotools
Eli Roberson (he/him) · @thatdnaguy
222 followers · 305 posts · Server genomic.socialDo I know anyone working with #nanostring #GeoMx? This is the first GeoMx dataset we've received.
There appear to be several packages for upfront QC and transfer to Seurat of the *processed* data in R. But I wasn't sure if there was anyone with recommendations for taking FASTQ to DCC. The instructions for installing and running the Nanostring pipeline don't seem very friendly to pure command-line interaction on a local server of in self-contained docker image.
#bioinformatics #genomics #nanostring #geomx
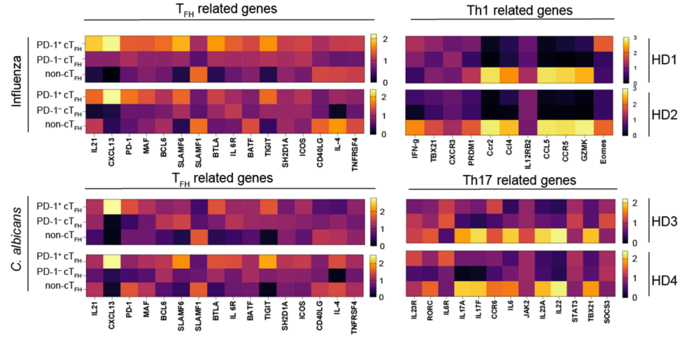
Antonino Cassotta · @ntoohm
4 followers · 7 posts · Server mas.toIn depth phenotyping by #Nanostring showed that Flu-specific but not C.alb-specific PD-1+ cTfh cells had a “GC Tfh-like” phenotype, with overexpression of IL21, CXCL13, and BCL6