Fatih Demir · @kabalak
57 followers · 116 posts · Server mstdn.scienceHmm, interesting questions: when doing in-vitro protease digests with recombinant protease, do I exclude self-cleavages of the protease from the putative cleavages as it might be rather an artefact of the excess protease addition? #Proteases #Ntermini
Fatih Demir · @kabalak
53 followers · 98 posts · Server mstdn.scienceAnyone else doing plasma proteomics with short nano gradients on an Ultimate3000 (<1h) and FAIMS/Exploris480 combined? I thought we had the golden rule of only 1 CV with shorter gradients lengths, but seems to be not true in terms of peptide-centric working people. #Ntermini #FAIMS #TeamMassSpec
#ntermini #faims #TeamMassSpec
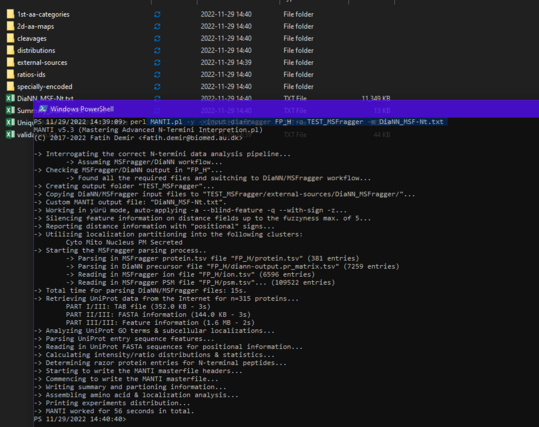
Fatih Demir · @kabalak
18 followers · 19 posts · Server mstdn.scienceAnother work in progress release of MANTI is out now! MANTI v5.3 (https://sourceforge.net/projects/manti/files/MANTI-v5.3.zip) enables you to analyze DiaNN/MSFragger-generated N-terminomics with the one-stop-shop approach of MANTI! To honor this, MANTI stands now for Mastering Advanced N-Termini Interpretation. It works for simplex data (only dimethyl-labelling currently) with the DIA_SpecLib_Quant workflow in MSFragger - currently, only DIA is supported for MSFragger, but more might be on the way. #MANTIv5.3 #Ntermini #proteomics
#mantiv5 #ntermini #proteomics