preLights · @preLights
595 followers · 34 posts · Server biologists.socialDiscovering the dynamic world of the EGFR-MAPK phosphoproteome. 🌐🔎
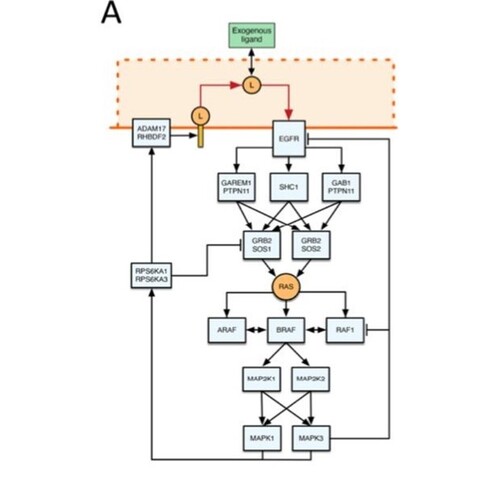
Feng, Sanford and colleagues #EMSLscience developed a multiplexed deep phosphoproteome profiling workflow unveiling 4500 protein sites exhibiting increased phosphorylation upon EGF stimulation.
Another great #preLight by Benjamin Maier 👉 https://prelights.biologists.com/highlights/a-phosphoproteomics-data-resource-for-systems-level-modeling-of-kinase-signaling-networks-v2/
#preprint # modelling #phosphoproteomics #signalling #bioinformatics #SystemsBiology
#emslscience #prelight #preprint #phosphoproteomics #signalling #bioinformatics #systemsbiology
Mol Syst Biol · @MolSystBiol
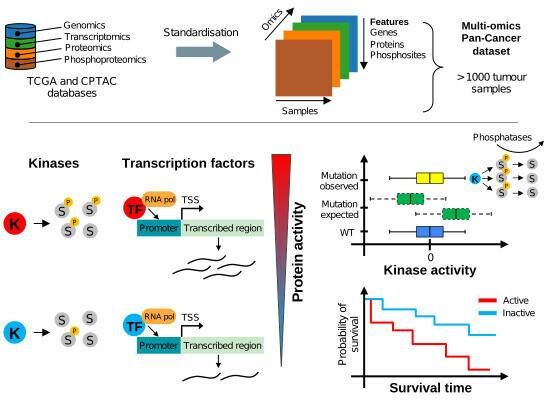
203 followers · 18 posts · Server sciencemastodon.comEstimation of activity changes for >500 kinases & transcription factors in >1,000 cancer samples & cell lines identifies drivers of signalling dysregulation & patient survival ➡️ bit.ly/3Jb9l5b
@pedrobeltrao IMSB ETH
@saezlab UniHeidelberg
@Epetsalaki EMBL-EBI
#CancerGenomics #Genomics #Phosphoproteomics #SystemsBiology
#Cancergenomics #genomics #phosphoproteomics #systemsbiology
Steve Baker · @Bakersm11
0 followers · 129 posts · Server theblower.au@edwardlau fantastic, #phosphoproteomics and #transcriptional profiling kinetics of #chromatin remodeling ?
#phosphoproteomics #transcriptional #chromatin
Steve Baker · @Bakersm11
0 followers · 117 posts · Server theblower.au@seemasainson @J_Cell_Sci @Dev_journal
#integrins, #transcription factors , #cadhedrins, #phosphoproteomics, #glycomics, #biofilm, #quorum receptors, #idiotype, #immnoglobulin subclass swithcing, #immunogenetics, #cytogenetics, #phylogenetics #metabolmics , #reverse vaccinology, etc.?
#integrins #transcription #cadhedrins #phosphoproteomics #glycomics #biofilm #quorum #idiotype #immnoglobulin #immunogenetics #cytogenetics #phylogenetics #metabolmics #reverse