Francisco Rodriguez-Sanchez · @frod_san
105 followers · 155 posts · Server ecoevo.socialRT @mvugt
#PLOSCompBio: Ten simple rules for working with other people’s code https://dx.plos.org/10.1371/journal.pcbi.1011031
CavalliLab · @giacomocavalli
148 followers · 50 posts · Server mastodon.social#PLOSCompBio: Phenotypic pliancy and the breakdown of epigenetic polycomb mechanisms https://dx.plos.org/10.1371/journal.pcbi.1010889
The authors call "phenotypic pliancy" the decreased phenotypic fidelity, resulting from dysregulation of specific cellular processes, allowing dysregulated cells to sustainably switch their phenotype in response to environmental changes. They claim that Polycomb dysregulation induces phenotypic pliancy and that this might have important consequences, notably in metastasis.
Luis M. Rocha · @lmrocha
168 followers · 173 posts · Server qoto.orgSo happy to finally see this collaboration with Rion Correia and @alainbarrat out. The distance backbone is a unique, algebraically-principled network subgraph that preserves all shortest paths. We were were excited to find out (with #sociopatterns and other data) that the backbones of #socialnetworks contain large amounts of redundant interactions that can be removed with very little impact on #communitystructure and #epidemic spread.
#complexsystems #dynamics
#Networkscience #Sparsification #complexnetworks
#PLOSCompBio:
#sparsification #networkscience #complexnetworks #ploscompbio #sociopatterns #socialnetworks #complexsystems #communitystructure #epidemic #dynamics
Mary · @mary
339 followers · 2512 posts · Server queer.af#paper #PLOSCompBio: A mathematical model of potassium homeostasis: Effect of feedforward and feedback controls https://dx.plos.org/10.1371/journal.pcbi.1010607
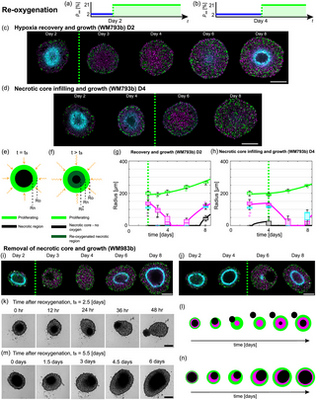
Mat Simpson · @profmjsimpson
59 followers · 184 posts · Server mathstodon.xyzRT @HaassLab@twitter.com
Hot off the press - exciting story, great collaboration!
@RyanMurphy42@twitter.com, @ProfMJSimpson@twitter.com, @QUT@twitter.com, @HaassLab@twitter.com, #FrazerInstUQ, @UQMedicine@twitter.com, @tri_australia@twitter.com, #PLOSCompBio: Growth and adaptation mechanisms of tumour spheroids with time-dependent oxygen availability https://dx.plos.org/10.1371/journal.pcbi.1010833
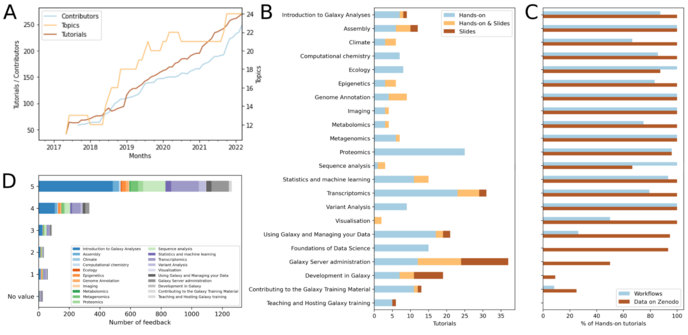
Bérénice Batut · @bebatut
43 followers · 4 posts · Server piaille.fr✨ The good news of the day 🤗
📰 Our paper "Galaxy #training: A powerful framework for teaching!" is now published #PLOSCompBio
➡️ https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1010752
❤️ Thanks to all @gtn & @galaxyproject ecosystem contributors, & to the reviewers for their valuable comments
CavalliLab · @giacomocavalli
111 followers · 13 posts · Server mastodon.social#PLOSCompBio: HiSV: A control-free method for structural variation detection from #HiC data https://dx.plos.org/10.1371/journal.pcbi.1010760
Martin Vinck · @martin_a_vinck
259 followers · 1 posts · Server neuromatch.socialVery happy that this new paper led by PhD student in our lab Jinke Liu is now finally out:
#PLOSCompBio: Improved visualization of high-dimensional data using the distance-of-distance transformation
Jinke shows here how in high-dimensional spaces tSNE and UMAP have a crowding problem due to scatter noise, which invades the cluster regions.
This problem can be solved by using a distance-of-distance transformations, where distance is now defined by similarity of distances to the local neighbourhoods of two points.
Attached an example of how this improves the tSNE embedding.
Jinke shows that this technique leads to improved visualization of high-dimensional neural representations during different network states, as well as image representations in convolutional neural networks.
Justin Stewart · @thecrobe
673 followers · 320 posts · Server ecoevo.socialRT @symbioticworld@twitter.com
#PLOSCompBio: How public can public goods be? Environmental context shapes the evolutionary ecology of partia ... https://dx.plos.org/10.1371/journal.pcbi.1010666
🐦🔗: https://twitter.com/symbioticworld/status/1605159995611533313
Dilek Koptekin · @dilekopter
22 followers · 8 posts · Server ecoevo.socialCheck out our new paper where we developed a new algorithm called CONGA that can genotype large deletions and duplications in low-coverage ancient genomes.
#PLOSCompBio: CONGA: Copy number variation genotyping in ancient genomes and low-coverage sequencing data https://dx.plos.org/10.1371/journal.pcbi.1010788
Francois Sabot · @francois_sabot
230 followers · 232 posts · Server genomic.socialRT @carlystrasser
Our paper finally came out in #PLOSCompBio! Ten simple rules for funding scientific #OpenSource software https://dx.plos.org/10.1371/journal.pcbi.1010627
Francois Sabot · @francois_sabot
189 followers · 147 posts · Server genomic.socialRT @edanchin
AvP: our method for robust phylogenetic detection of horizontal gene transfers at high-throughput freshly published in #PLOSCompBio: https://dx.plos.org/10.1371/journal.pcbi.1010686 hope AvP will accelerate HGT identification in many new genomes !
Francois Sabot · @francois_sabot
189 followers · 147 posts · Server genomic.socialRT @PNDBiodiv
#PLOSCompBio: Ten simple rules to cultivate belonging in collaborative data science research teams https://dx.plos.org/10.1371/journal.pcbi.1010567