Science Magazine :press: · @ScienceMagazine
391 followers · 52 posts · Server press.coopScientists present a computational strategy that mimics gene duplication to predict new #ProteinProteinInteractions—enabling exploration of uncharted regions of the protein sequence space and design of new interacting protein pairs. https://scim.ag/2b8 @scisignal #press
#proteinproteininteractions #press
Mol Syst Biol · @MolSystBiol
229 followers · 36 posts · Server sciencemastodon.comA new proteomic workflow establishing interdependencies between protein #phosphorylation & complex formation is applied to the signal integrator & Hippo pathway effector YAP1 ➡️ http://bit.ly/3mJ4W0t
Matthias Gstaiger @eth_en
#proteomics #interactome #ProteinProteinInteractions #CellSignaling
#phosphorylation #proteomics #interactome #proteinproteininteractions #cellsignaling
Mol Syst Biol · @MolSystBiol
229 followers · 35 posts · Server sciencemastodon.comNews &Views by
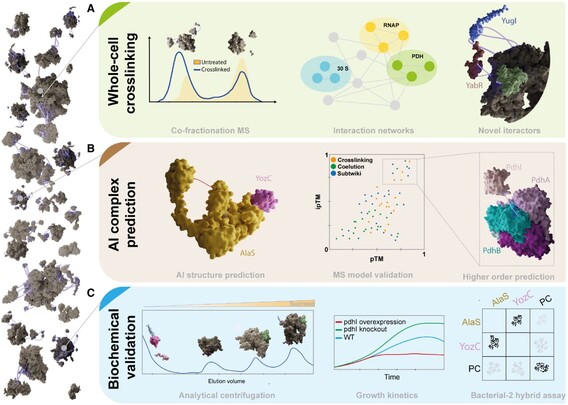
P Kastritis on the recent study from the Rappsilber lab identifying bacterial PPIs in their cellular context by combining whole-cell crosslinking, co-fractionation #massspectrometry & AI-based PPI structure prediction ➡️http://bit.ly/3FfcIVY
#proteomics #ProteinProteinInteractions #SystemsBiology #Alphafold
#massspectrometry #proteomics #proteinproteininteractions #systemsbiology #AlphaFold
Mol Syst Biol · @MolSystBiol
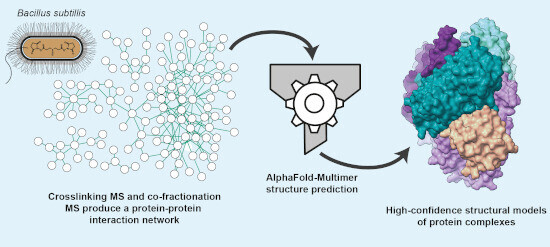
226 followers · 30 posts · Server sciencemastodon.comAn integrative approach using crosslinking #massspectrometry (MS), co-fractionation MS & #Alphafold-Multimer discovers new protein complexes & their topologies in B.subtillis ➡️ http://bit.ly/3kpjocU
from Juri Rappsilber (TU Berlin), Jörg Stülke (Uni Goettingen)
#proteomics #SystemsBiology #ProteinProteinInteractions
#massspectrometry #AlphaFold #proteomics #systemsbiology #proteinproteininteractions
Mol Syst Biol · @MolSystBiol
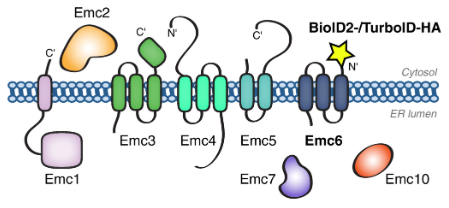
201 followers · 17 posts · Server sciencemastodon.comABOLISH: an optimised biotin-ligation method for discovering stable & transient #ProteinProteinInteractions in yeast ➡️ bit.ly/3ktzuBT
from Maya Schuldiner's lab, Weizmann Institute
#SystemsBiology #Scerevisiae
#proteinproteininteractions #systemsbiology #scerevisiae