Pete Carlton · @pmcarlton
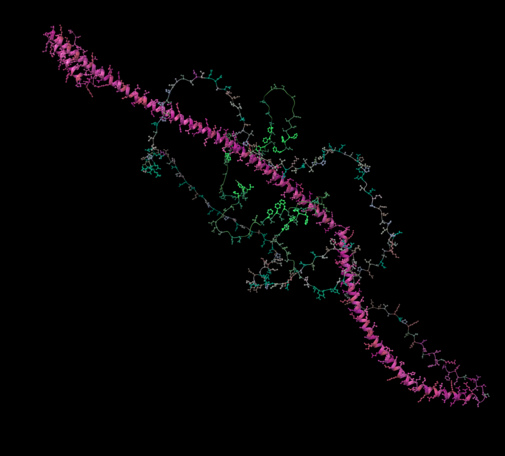
120 followers · 155 posts · Server fediscience.orgI am still finding interesting things to do with protein #embeddings — now coloring #AlphaFold models by the 3-component UMAP reduction of the per-residue embedding. Look at those bright Phe residues in the disordered region — somehow #protT5 encodes them as "different" than the others.
In my experience so far these protein language models are uncannily able to highlight the same regions of the protein that I'm interested in already.
(#ChimeraX, using a sequence coloring format file)
#chimerax #prott5 #alphafold #embeddings
Last updated 3 years ago