Teresita Porter 🙋🏻♀️ · @DNAdataPhile
147 followers · 548 posts · Server ecoevo.social**A global baseline for qPCR-determined antimicrobial resistance gene prevalence across environments**
https://www.sciencedirect.com/science/article/pii/S0160412023003574?via%3Dihub
#AMR #AntiMicrobialResistance #qPCR #EnvironmentalMonitoring
#AMR #antimicrobialresistance #qPCR #environmentalmonitoring
RoedigerRG · @RoedigerRG
13 followers · 27 posts · Server social.anoxinon.deNext week starts the 10th Gene Quantification Event #GQ2023 in Freising. Focus Topics are
Sars-Cov-2, Spatial- #Transcriptomics, #LiquidBiopsy & #CNA, Multi-#Omics #Biomarkers
Looking forward to this. We will present some research about #machinelearning on qPCR data (https://joss.theoj.org/papers/10.21105/joss.04407).
#gq2023 #transcriptomics #liquidbiopsy #cna #omics #biomarkers #machinelearning #qPCR #dpcr #ngs
RoedigerRG · @RoedigerRG
4 followers · 13 posts · Server social.anoxinon.de#GQ2023 — 10th Gene Quantification Event in #Freising is great because you can learn for example about #NGS, #qPCR, #dPCR, #biomarkers, Spatial-#Transcriptomics, #LiquidBiopsy & Circulating Nucleic Acids by great scientists like Jim #Huggett (#dMIQE), Stephen #Bustin (https://en.wikipedia.org/wiki/Stephen_Bustin), Michael W. #Pfaffl (https://scholar.google.de/citations?user=mYAWj3gAAAAJ&hl=de) or Jo #Vandesompele (famous for #qBase, #geNorm, ...) https://gene-quantification-2023.events/
#gq2023 #freising #ngs #qPCR #dpcr #biomarkers #transcriptomics #liquidbiopsy #huggett #dmiqe #Bustin #pfaffl #vandesompele #qbase #genorm
Anna MacDonald · @dr_annam
936 followers · 118 posts · Server ecoevo.socialEnvironmental DNA lets us detect species, but DNA moves & persists in the environment, so a DNA detection may not represent a local, living individual. Our #NewPaper (led by Léonie Suter) uses different DNA fragment lengths to distinguish recently-shed from older krill eDNA
Read the paper here: https://onlinelibrary.wiley.com/doi/full/10.1002/edn3.394
AusAntarctic news summary here: https://www.antarctica.gov.au/news/2023/dna-detector-exposes-how-antarctic-krill-use-habitat/
#newpaper #eDNA #environmentalDNA #qPCR #Antarctic #krill
Anna MacDonald · @dr_annam
936 followers · 118 posts · Server ecoevo.socialEnvironmental DNA lets us detect species, but DNA moves & persists in the environment, so a DNA detection may not represent a local, living individual. Our new paper (led by Léonie Suter) uses different DNA fragment lengths to distinguish recently-shed from older krill eDNA
Read the paper here: https://onlinelibrary.wiley.com/doi/full/10.1002/edn3.394
AusAntarctic news summary here: https://www.antarctica.gov.au/news/2023/dna-detector-exposes-how-antarctic-krill-use-habitat/
#eDNA #environmentalDNA #qPCR #Antarctic #krill
Jessica Rieder · @jess_rieder
45 followers · 70 posts · Server ecoevo.social📝The development of field-ready #qPCR assays for the #monitoring and detection of 3 #aquatic #pathogens, comparing 2 different drying methods. Combine with portable qPCR machines or lateral flow strips for on-site detection. https://doi.org/10.1016/j.mimet.2022.106594
#qPCR #monitoring #aquatic #Pathogens
devSJR :python: :rstats: · @devSJR
124 followers · 117 posts · Server fosstodon.orgSome colleagues and I have been working for a long time on a review of R packages for the analysis of #qPCR experiments, #dPCR experiments and melting curves. What we can find is that there are a lot of packages and we will probably overlook some. I did my last search with https://rdrr.io/ and discovered a lot more.
Helmut Bürgmann · @HelmutBuergmann
32 followers · 9 posts · Server mstdn.scienceHere is my #introduction - I am a #microbial #ecologist with Eawag: #Swiss Federal Institute of #Aquatic #Science and Technology in the Surface Waters department. Interested in the role of #microbes in #biogeochemistry of #lake, #river, #sediment & #wastewater. Also #research on #AntibioticResistance in the #environment.
We use #metagenomics, #metatranscriptomics #qPCR and good old-fashioned #microbiology.
#MicrobialEcology
#EnvironmentalMicrobiology
#AMR #Antibiotic
#methane #nitrogen
#introduction #microbial #ecologist #Swiss #aquatic #Science #microbes #biogeochemistry #lake #river #sediment #wastewater #research #antibioticresistance #environment #Metagenomics #metatranscriptomics #qPCR #microbiology #microbialecology #environmentalmicrobiology #AMR #antibiotic #methane #nitrogen
devSJR :python: :rstats: · @devSJR
124 followers · 117 posts · Server fosstodon.org@chanceyork
Mainly bioinformatics with #rstats and #RKWard beeing my friends. Sometimes #Python helps to solve problems. #datatable (the #SQL of #R) is what I often use. My raw data originate from methods like #qPCR, #dPCR, #NGS and image data of patient data (count data, time series) keep me busy. Lately I worked with text data in my work related to #forensics.
#rstats #rkward #python #datatable #sql #r #qPCR #dpcR #ngs #forensics
Dr Biswapriya Misra · @BiswapriyaMisra
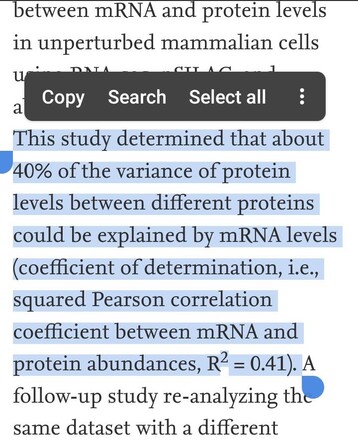
105 followers · 30 posts · Server fosstodon.orgIn 2011, Schwanhäusser, et al. (Nature, 473(7347) measured #absolute #protein & #mRNA levels for 5000 genes in #mamalian cells and informed us 👇
"Protein & mRNA levels correlates 0.41"
Yet, #Reviewer2 demands:
1. #Proteomics validated by #qPCR
2. #RNAseq based #biomarkers
#absolute #protein #mrna #mamalian #Reviewer2 #proteomics #qPCR #rnaseq #biomarkers
devSJR :python: :rstats: · @devSJR
124 followers · 117 posts · Server fosstodon.orgRecently I used https://r-pkg.org/ (
#METACRAN: Search and browse all #CRAN/#R packages). This was a good thing to do, since I discovered R packages related to #qPCR that I haven't heard of before. Since several authors and I are writing a review about #dPCR, qPCR and #meltingcurveanalysis this is good not to forget anybody.
#METACRAN #cran #qPCR #dpcR #meltingcurveanalysis #rstats