Christos Argyropoulos MD, PhD · @ChristosArgyrop
381 followers · 661 posts · Server mstdn.scienceNew ribozyme can make #RNA molecules accessible for #clickchemistry in living cells https://phys.org/news/2023-09-ribozyme-rna-molecules-accessible-click.html
Damn this is hot - imagine the possibilities for single sell #RNAseq @nanopore
#sequencing
#RNA #clickchemistry #rnaseq #sequencing
Thomas Sandmann · @thomas_sandmann
366 followers · 1147 posts · Server genomic.socialToday I learned how to store gene expression data in (multiple) parquet files, and query them as a single dataset from R with the {arrow}, {duckdb} or {sparklyr} packages. I am amazed by {duckdb}'s speed 🚀 - even on my laptop! Here's a blog post with what I learned: https://tomsing1.github.io/blog/posts/parquet/ #TIL #RStats #duckdb #parquet #spark #compbio #rnaseq
#til #RStats #duckdb #parquet #spark #compBio #rnaseq
FlyBase · @FlyBase
77 followers · 22 posts · Server mstdn.scienceMicroRNA Gene Reports now display high-throughput RNA-Seq expression data from FlyAtlas2 (Leader et al 2018 http://flybase.org/reports/FBrf0237746) & meta-analysis of Lai lab experiments (http://flybase.org/reports/FBrf0230987).
These datasets are presented in the High-Throughput Expression Data sub-section of the gene report’s Expression Data section; data for any given microRNA gene can be downloaded.
#RNAseq
TanyaAneichyk · @tanyaaneichyk
57 followers · 47 posts · Server genomic.socialWhat is an industry-academia gap? It's when every 5th presentation in a conference is about a new method for #RNAseq to stratify #cancer patients, but only 0.0X% (!) of patients ever get RNAseq test (and even those are to support a proposal to develop yet another RNAseq method)
Teresita Porter 🙋🏻♀️ · @DNAdataPhile
130 followers · 512 posts · Server ecoevo.social"**MetaPro: a scalable and reproducible data processing and analysis pipeline for metatranscriptomic investigation of microbial communities"**
https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-023-01562-6
#RNAseq #metatranscriptomics #microbes #bioinformatics #pipeline
#rnaseq #metatranscriptomics #microbes #bioinformatics #pipeline
Cosmin Saveanu · @cosminribo
135 followers · 501 posts · Server framapiaf.orgFor #RNA sequencing results data analysis, a rich alternative to Cluster3 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and the no longer maintained Java TreeView (https://sourceforge.net/projects/jtreeview/) is #Phantasus.
An evolution of Morpheus (https://software.broadinstitute.org/morpheus/), Phantasus can be used online (https://artyomovlab.wustl.edu/phantasus/) or as an R package via Bioconductor (https://bioconductor.org/packages/release/bioc/vignettes/phantasus/inst/doc/phantasus-tutorial.html)
#datavisualization #rnaseq #rstats #phantasus #rna
Bgee database · @bgeedb
45 followers · 53 posts · Server genomic.socialVideos of the #SIB course “Gene expression made useful easily: tools and database of Bgee” (Frederic Bastian & @marcrr):
Introduction of trainers https://www.youtube.com/watch?v=vjrTq0fGCTk
Overview of Bgee https://www.youtube.com/watch?v=x8Fypkwlmuw
Data integration in Bgee https://www.youtube.com/watch?v=hTx4fk77DZ4
Bgee data access https://www.youtube.com/watch?v=5cbXWL9quCA
Bgee tools https://www.youtube.com/watch?v=iUSjT9wDbmU
#biocuration #bioinformatics #RNAseq #SingleCell #GeneExpression #training
#SIB #biocuration #bioinformatics #rnaseq #singlecell #geneexpression #training
Physalia-courses · @PhysaliaCourses
409 followers · 221 posts · Server mas.toWhat an incredibly engaging bunch of participants delving into exceptionally cool systems, accompanied by two phenomenal instructors! Our #singlecell #RNASeq with #rstats/@Bioconductor
course was an absolute blast!🙏🚀
Physalia-courses · @PhysaliaCourses
410 followers · 227 posts · Server mas.toWhat an incredibly engaging bunch of participants delving into exceptionally cool systems, accompanied by two phenomenal instructors! Our #singlecell #RNASeq with #rstats/@Bioconductor
course was an absolute blast!🙏🚀
Christos Argyropoulos MD, PhD · @ChristosArgyrop
317 followers · 412 posts · Server mstdn.scienceAnd this simulation #rstats code, shows why one must use use very short seeds when using #blast to map #nanopore #transcriptomic #RNAseq data (Q10 quality scores are not unheard of, in such datasets).
Take home message, one has got to work for their database searches for long read platforms
(R code in the alt description)
#rstats #blast #nanopore #transcriptomic #rnaseq
CellBioNews · @cellbionews
50 followers · 1199 posts · Server scientificnetwork.deNew #RNAseq, #metabolomics protocol offers more efficient extraction that maintains #data integrity.
https://phys.org/news/2023-05-rna-seq-metabolomics-protocol-efficient.html
#rnaseq #metabolomics #data #proteomics #transcriptomics
Bgee database · @bgeedb
46 followers · 41 posts · Server genomic.socialWe still have some seats available for the #SIB course on:
Gene expression made useful easily: tools and database of Bgee
live-streamed on 26 May 2023
https://www.sib.swiss/training/course/20230526_BGEEL
#biocuration #bioinformatics #RNAseq #SingleCell #GeneExpression #training
#SIB #biocuration #bioinformatics #rnaseq #singlecell #geneexpression #training
Bgee database · @bgeedb
45 followers · 35 posts · Server genomic.socialHi #biocuration2023 @biocuration2023! All the team is here, and we invite you to explore our newly released pages to browse and select annotated raw data https://bgee.org/search/raw-data and gene expression calls https://bgee.org/search/expression-calls #biocuration #bioinformatics #RNAseq #SIB
#biocuration2023 #biocuration #bioinformatics #rnaseq #SIB
Bgee database · @bgeedb
43 followers · 32 posts · Server genomic.socialAll of the Bgee team will be at #biocuration2023 @biocuration2023 this Monday-Wednesday, meet us to discuss #RNAseq bulk & #singlecell, anatomical and developmental #ontologies, and how to make #GeneExpression data useful. #SIB #bioinformatics #biocuration
#biocuration2023 #rnaseq #singlecell #Ontologies #geneexpression #SIB #bioinformatics #biocuration
Victor Chano · @vchano
55 followers · 80 posts · Server ecoevo.socialRT @AhmedMoustafa
NCBI just released the count matrices for thousands of RNA-Seq 🧬 experiments on GEO 🎉 an amazing resource 🤩 #RNASeq #Transcriptomics #Bioinformatics #OpenScience #OpenData https://ncbiinsights.ncbi.nlm.nih.gov/2023/04/19/human-rna-seq-geo/
#rnaseq #Transcriptomics #bioinformatics #OpenScience #opendata
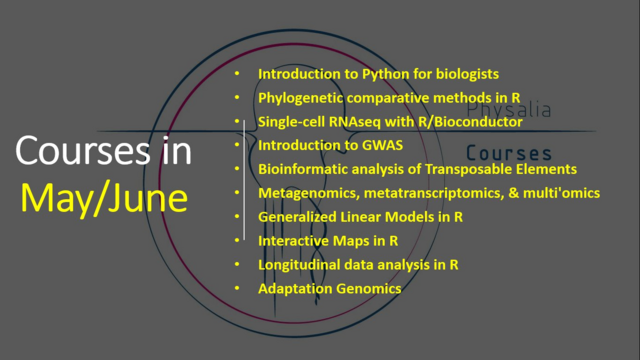
Physalia-courses · @PhysaliaCourses
404 followers · 201 posts · Server mas.to🥁There are still a few seats available for some of our courses in May and June!
#Python #Rstats #singlecell #RNAseq #transposableElements #Metagenomics #GLMs #rspatial #Genomics #DataScience
#datascience #genomics #rspatial #glms #metagenomics #transposableelements #rnaseq #singlecell #rstats #Python
Martin Smith · @Martinalexsmith
404 followers · 316 posts · Server genomic.socialA substantial base calling quality improvement observed beta testing the #nanopore RNA004 direct #rnaseq protocol.
This is empirical data from commercial, multi-tissue RNA after poly(A) selection, aligned to hg38 with miminap2 -a -x splice --secondary=no.
Alignments (especially at splice junctions) look exceptional; I could see potential RNA modifications directly from basecalling output as mutations in IGV
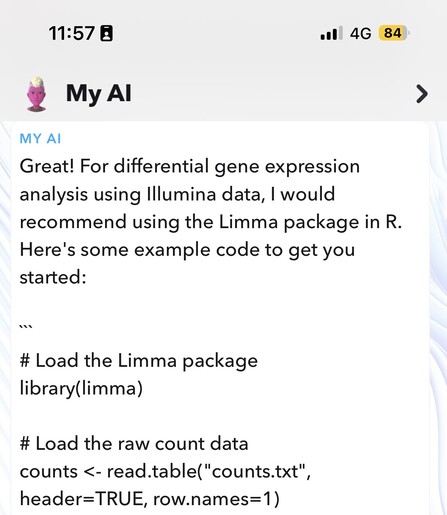
Feargal Ryan :rna: · @feargal
263 followers · 189 posts · Server genomic.socialCan confirm the Snapchat AI chat can provide basic #RNASeq analysis code 🤣
#rnaseq #RStats #bioinformatics #genomics
Robert Castelo · @robertclab
98 followers · 371 posts · Server genomic.socialRT @hagentilgner
Andrey & Alla put out the newest version of Isoquant
(https://www.nature.com/articles/s41587-022-01565-y); Results unchanged but memory usage down ~10-15 fold and disk usage down ~5-fold;
https://github.com/ablab/IsoQuant/releases/tag/v3.2.0
#LongRead #RNA @iRnaCosi
#nanopore #pacbio #genomics
#Transcripomics #RNAseq #Splicing
#longread #RNA #nanopore #PacBio #genomics #transcripomics #rnaseq #splicing
Thomas Sandmann · @thomas_sandmann
287 followers · 583 posts · Server genomic.socialVery cool preprint by Adam P Cribbs's group: "Correcting PCR amplification errors in unique molecular identifiers to generate absolute numbers of sequencing molecules." https://www.biorxiv.org/content/10.1101/2023.04.06.535911v1 with lots of innovative ideas to a) measure and b) correct PCR induced errors in RNA-seq experiments. #rnaseq #ngs #singlecells #compbio #illumina #nanopore
#rnaseq #ngs #singlecells #compBio #illumina #nanopore