Daniel Caron · @carondanielp
32 followers · 37 posts · Server genomic.socialSo excited to share MMoCHi: a tool I've built for multi-modal cell type classification for CITE-seq data
The pre-print can be found here:
https://www.biorxiv.org/content/10.1101/2023.07.06.547944v1
And we've put it out on #GitHub so that you can try it out on your own data:
https://mmochi.readthedocs.io
#scRNAseq #scRNA #CITEseq #sequencing #analysis #ColumbiaUniversity #immunology
#github #scrnaseq #scRNA #citeseq #sequencing #analysis #columbiauniversity #immunology
Mito · @mito
0 followers · 4 posts · Server genomic.socialThomas Sandmann · @thomas_sandmann
280 followers · 489 posts · Server genomic.socialJust read the incredible preprint "A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain" by Yao et al https://www.biorxiv.org/content/10.1101/2023.03.06.531121v1. An atlas of the whole mouse brain, with single-cell RNA-seq and spatial (MERFISH) data from > 4 million high-quality cells. The authors describe 5,000 distinguishable "clusters", the most corresponding to neurons with unique patterns of expression of ~ 500 transcription factors.A landmark paper. #scRNA #allen #spatial #neuroscience
#scRNA #allen #Spatial #neuroscience
António Domingues · @keyboardpipette
62 followers · 154 posts · Server genomic.social@haojiawu @biorxivpreprint this is very interesting. I have been telling people that one of the main reasons for using #RStats instead of #python for gene expression analysis, is the lack of methods such as #DESeq2 or #limma as python libraries. This might tip the balance for a lot of people.
Of course there are many reasons to prefer R, the excellent #Bioconductor ecosystem one of them, and in fairness, for #scRNA analysis python has very strong ecosystem and community.
#RStats #python #deseq2 #limma #Bioconductor #scRNA
Casey Greene · @greenescientist
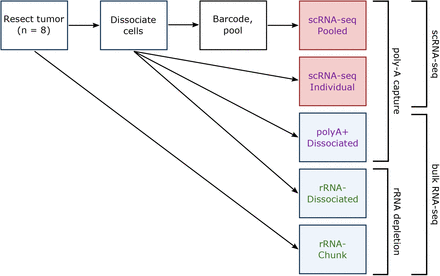
1022 followers · 79 posts · Server genomic.socialI'm excited about this new #genomics #scRNA #rnaseq preprint from @ariel_hippen! It's a molecular biology deep dive into what happens when you dissociate and single-cell sequence a tissue with the goal of performing deconvolution of bulk tissue.
Some key bits: there are dissociation effects, RNA capture/depletion effects, and potentially some microfluidics effects with the #10X platform. Also, deconvolution algorithms are variably robust to these things.
See more:
https://www.biorxiv.org/content/10.1101/2022.12.04.519045v1
Martin Fahrenberger · @FahrenbergerM
0 followers · 1 posts · Server genomic.socialHi I'm Martin,
a 4th year PhD-student in #bioinformatics working on #scRNA-seq methods development.
I work in Arndt von Haeseler's lab at the Max Perutz Labs, part of the Vienna BioCenter.
Follow me for updates on my research and the occasional uninformed joke about politics.
#introduction
#introduction #scRNA #bioinformatics
Christian Ray · @jjray
208 followers · 106 posts · Server social.tchncs.deBy way of #introduction for the great migration: I am a builder of models: computational (#ml, #machinelearning), mathematical, and scale models (#hoscale #scalemodel). Working as a computational biologist / bioinformaticist for ExosomeDx. (#multiomics #scRNA-seq #dimensionality #bioinformatics #computationalbiology #compbio) Follower of and/or interested in #osint, science history, nuclear power, #trains, pretty birds, canyon hikes, and scientific ethics.
#trains #osint #compBio #computationalbiology #bioinformatics #dimensionality #scRNA #multiomics #scalemodel #hoscale #machinelearning #ml #introduction