Knight ADRC · @KnightADRC
54 followers · 553 posts · Server universeodon.comUsing #snRNAseq to understand the #genetic architecture of rare & common variants for #AlzheimerDisease - https://idp.nature.com/authorize?response_type=cookie&client_id=grover&redirect_uri=https%3A%2F%2Fwww.nature.com%2Farticles%2Fs41467-023-37437-5
#snrnaseq #genetic #alzheimerdisease
Robert Castelo · @robertclab
92 followers · 238 posts · Server genomic.socialRT @lahuuki
Hot of the pre-print press! 🔥 Our latest work #spatialDLPFC pairs #snRNAseq and #Visium spatial transcriptomic data in the human #DLPFC building a neuroanatomical atlas of this critical brain region 🧠
@LieberInstitute @10xGenomics #scitwitter
📰 https://doi.org/10.1101/2023.02.15.528722
#spatialdlpfc #snrnaseq #visium #dlpfc #scitwitter
Augusto Escalante · @Augusto_ER
21 followers · 15 posts · Server neuromatch.social#Introduction time it is!
Hi everyone, just moved instances following really good advice (thanks @jonny ! 😉).
I'm a postdoc at the Instituto de Neurociencias (Alicante, Spain) working on the neural circuits of itch sensation at the level of the spinal cord dorsal horns and how these circuits are affected during chronic diseases cursing with pruritus (a major unmet medical need, for those of you far from this field). I'm currently analizing a #snRNAseq dataset I generated in a mouse model of chronic itch which we published a couple years ago from my work at the Max Planck Institute for Neurobiology (Munich, Germany) together with my then supervisor Rüdiger Klein. The aim is to identify the specific subpopulation of neurons responsible for the madly intense scratching phenotype.
Very interested also in behavioral analysis, we are trying to identify specific motor patterns in chronic itch mouse models using #deeplearning #SLEAP pose estimation and behavioral motif classification.
My background in neurodevelopmental biology, specifically axon guidance, also pushes me towards analyzing the developmental origin of spinal neurons populations in order to understand the emergence of these circuits during embryonic stages with the idea that knowing where we come from we might better try to fix broken things!
See you all around!!
#introduction #snrnaseq #deeplearning #sleap
Robert Castelo · @robertclab
75 followers · 95 posts · Server genomic.socialRT @lpachter
In a new preprint w/@kreldjarn, @DelaneyKSull, @GuillaumOleSan & @pmelsted we address a shortcoming in current approaches to quantifying single-nucleus RNA-seq: https://www.biorxiv.org/content/10.1101/2022.12.02.518832v1
tl;dr care has to be taken in quantifying nascent vs. mature transcripts. #snRNAseq #scRNAseq🧵
Anshul Kundaje · @akundaje
1282 followers · 469 posts · Server genomic.socialRT @lpachter@twitter.com
In a new preprint w/@kreldjarn, @DelaneyKSull@twitter.com, @GuillaumOleSan@twitter.com & @pmelsted@twitter.com we address a shortcoming in current approaches to quantifying single-nucleus RNA-seq: https://www.biorxiv.org/content/10.1101/2022.12.02.518832v1
tl;dr care has to be taken in quantifying nascent vs. mature transcripts. #snRNAseq #scRNAseq🧵
Marcel Schulz · @marcelschulz
91 followers · 201 posts · Server genomic.socialRT @lpachter
In a new preprint w/@kreldjarn, @DelaneyKSull, @GuillaumOleSan & @pmelsted we address a shortcoming in current approaches to quantifying single-nucleus RNA-seq: https://www.biorxiv.org/content/10.1101/2022.12.02.518832v1
tl;dr care has to be taken in quantifying nascent vs. mature transcripts. #snRNAseq #scRNAseq🧵
Yen-Chung Chen · @ycchen_tw
41 followers · 40 posts · Server drosophila.socialRT @lpachter@twitter.com
In a new preprint w/@kreldjarn, @DelaneyKSull@twitter.com, @GuillaumOleSan@twitter.com & @pmelsted@twitter.com we address a shortcoming in current approaches to quantifying single-nucleus RNA-seq: https://www.biorxiv.org/content/10.1101/2022.12.02.518832v1
tl;dr care has to be taken in quantifying nascent vs. mature transcripts. #snRNAseq #scRNAseq🧵
Prathyusha Konda · @prats
176 followers · 181 posts · Server fediscience.orgRT @lpachter@twitter.com
In a new preprint w/@kreldjarn, @DelaneyKSull@twitter.com, @GuillaumOleSan@twitter.com & @pmelsted@twitter.com we address a shortcoming in current approaches to quantifying single-nucleus RNA-seq: https://www.biorxiv.org/content/10.1101/2022.12.02.518832v1
tl;dr care has to be taken in quantifying nascent vs. mature transcripts. #snRNAseq #scRNAseq🧵
Posnien Lab · @posnienlab
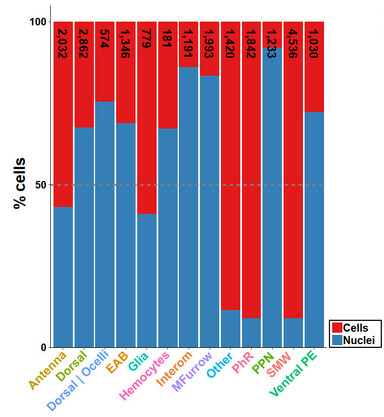
47 followers · 27 posts · Server fediscience.orgConclusion: For most applications, isolation of single nuclei followed by #snRNAseq is best choice if material is limited (e.g. early #development, tissue needs to be collected in the wild). Moreover, nuclei isolation protocol is much shorter and thus better reproducible. 7/8
Posnien Lab · @posnienlab
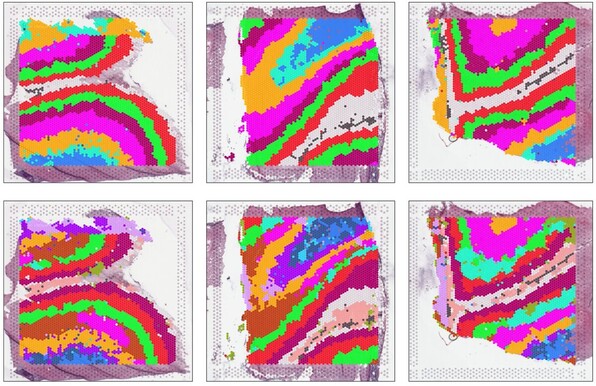
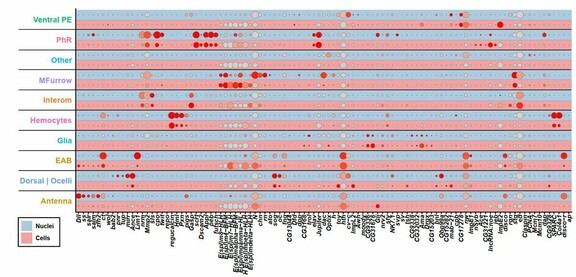
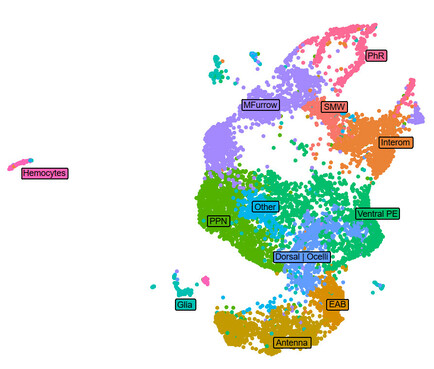
47 followers · 26 posts · Server fediscience.orgSome cell types (e.g. second mitotic wave) were more efficiently identified in #snRNAseq data and generally, we observed different contributions of both datasets to different cell types when we integrated both datasets. 6/8
Posnien Lab · @posnienlab
47 followers · 25 posts · Server fediscience.orgPosnien Lab · @posnienlab
47 followers · 24 posts · Server fediscience.orgA protocol based on the use of multiple RNase inhibitors was most efficient for fresh and snap frozen discs (protocol based on https://dx.doi.org/10.17504/protocols.io.veae3ae) and subsequent #snRNAseq revealed most relevant cell types. 4/8
Ludovic S Mure · @ludovicSmure
60 followers · 25 posts · Server mas.to#scRNAseq vs #snRNAseq: different cell types enrichment but correlated gene expression profiles (at least in the rabbit retina)
https://www.biorxiv.org/content/10.1101/2022.11.07.515504v1
Ruslan Rust · @ruslan
86 followers · 23 posts · Server mas.toHey #snRNAseq experts: We plan to transplant human cells in a mouse brain and sort+sequence the grafted nuclei 1mo after transplantation.
There are many protocols around, Which procedures can you recommend? Is there anything to watch out for? Please retweet ☺️🙏