Ben Neely · @neely
277 followers · 415 posts · Server fediscience.orgI am listening to this great lecture by Michelle Heck, and it made me think about some other great lectures I've missed (I still have a list from lockdown). One big one I haven't tracked down is the Jimmy Eng talk at HUPO 2020 (I think @pwilmart recommended it at the time). But... I have no idea where this video is! Any help from the masses, and sorry to be lazy.
Ben Neely · @neely
275 followers · 381 posts · Server fediscience.orgCheck me please #teamProteome: “quant” at the protein level seems normally distributed across most sample types (high and low abundance tails), but adding depth (new MS) doesn’t preserve the low abundance tail.
Is the drop off tail biologically “real”?
Ex. blood/secretome from Anderson^2 to Uhlén.
Ben Neely · @neely
267 followers · 339 posts · Server fediscience.orgI’m the late 00’s I was enamored with AFFF-MALS (or DLS), and am always surprised I am not hearing about folks using it upstream of proteomics. Could definitely work (and did for me in 2008) for EVs and likely work for some organelles and even some cell types (I don’t actually know the upper limit on size range).
Is it something Wyatt did or…?
#teammassspec #teamproteome #Proteomics
Ben Neely · @neely
265 followers · 320 posts · Server fediscience.orgWant to thank everyone here for some stellar discussion on SAAVs and searching.
Follow up: is there a SAAV-only open search option? So open-search but constrain fitting delta mass unknowns to AAs only (no funky ptms). I am off to poke around in #FragPipe but figured the crowd might know better.
#teamproteome #teammassspec #fragpipe
Ben Neely · @neely
261 followers · 285 posts · Server fediscience.orgHonestly, maybe this is like when I (personally) found mass spec twitter c2018: it was @educhicano pastelbiosciences and @UCDProteomics dropping links. The #teamproteome contingent is much stronger here now with a ton of heavy hitters.
I still have issues with the timeline, but at least I can get my nerd on without a limit.
NorSivaeb · @RonBeavis
123 followers · 667 posts · Server mastodon.socialDo you include Y-chromosome linked protein sequences in the analysis of samples that cannot contain them (e.g. female tissue or cell lines like HeLa, HEK-293 or MCF-7)? #teamProteome
NorSivaeb · @RonBeavis
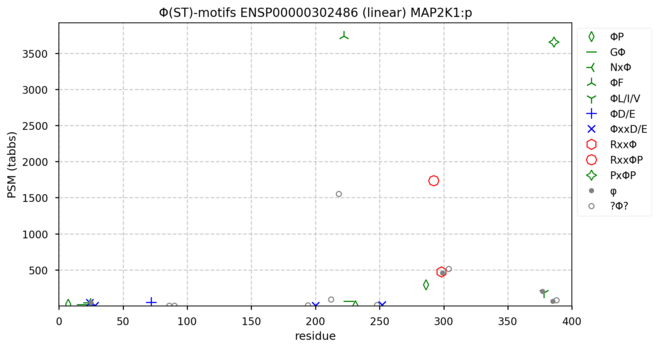
123 followers · 665 posts · Server mastodon.socialΦ(ST) Motif diagrams for human & mouse mitogen-activated protein kinase kinase 1―MAP2K1:p―aka MEK1, MAPKK1 & PRKMK1. Three major, conserved S/T phosphodomains anchored by ΦF, RxxΦP and PxΦP acceptor motifs. #teamProteome
NorSivaeb · @RonBeavis
123 followers · 657 posts · Server mastodon.socialProbably worth a look if you are interested in this sort of thing. #teamProteome
https://pubs.acs.org/doi/10.1021/acs.jproteome.2c00388
NorSivaeb · @RonBeavis
123 followers · 654 posts · Server mastodon.socialΦ(ST) Motif diagrams for human & mouse ubiquitin specific peptidase 5―USP5:p―has 3 discrete phosphodomains involving conserved acceptor site motifs. #teamProteome
NorSivaeb · @RonBeavis
122 followers · 649 posts · Server mastodon.socialDo you think you have as intuitive a feel for data when it is plotted with a log scale compared to the same data plotted with a linear scale? #teamProteome
NorSivaeb · @RonBeavis
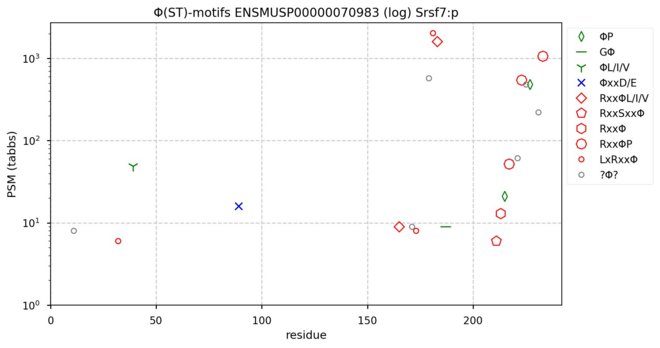
122 followers · 643 posts · Server mastodon.socialΦ(ST)-motifs diagram for mouse serine/arginine-rich splicing factor 7―if I ever wrote a monograph explaining the mechanisms that give rise to the apparent complexity of ST phosphorylation patterns, this would be Fig. 1. #teamProteome
NorSivaeb · @RonBeavis
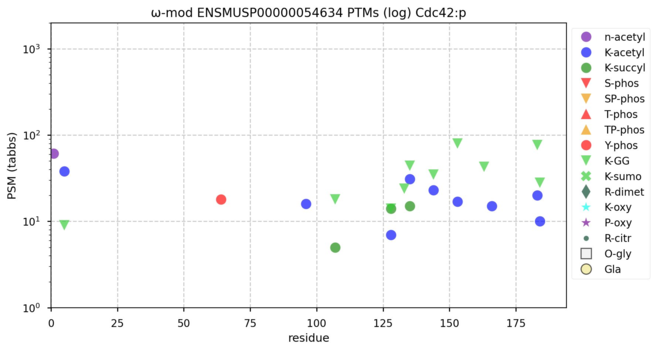
122 followers · 636 posts · Server mastodon.socialModification diagrams for mouse cell division cycle 42, CDC42 binding protein kinase alpha & beta―CDC42:p, CDC42BPA:p, CDC42BPB:p―a small GTPase & 2 kinases that bind it. The kinase genes are paralogues & their protein PTM patterns are proteologues. #teamProteome
NorSivaeb · @RonBeavis
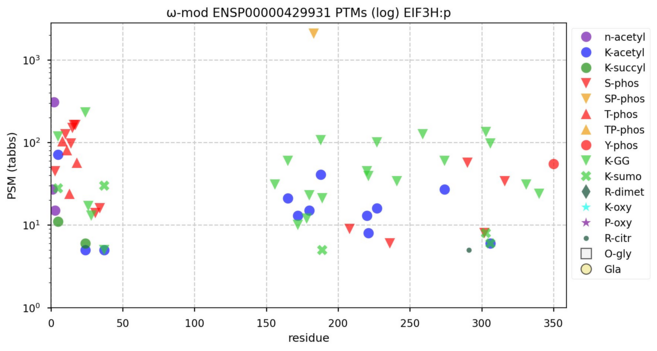
122 followers · 635 posts · Server mastodon.socialModification & W-S diagram for mouse eukaryotic translation initiation factor 3 subunit H―EIF3H:p―a component of the 43S preinitiation complex with several domains that can be easily differentiated by their PTM patterns. #teamProteome
NorSivaeb · @RonBeavis
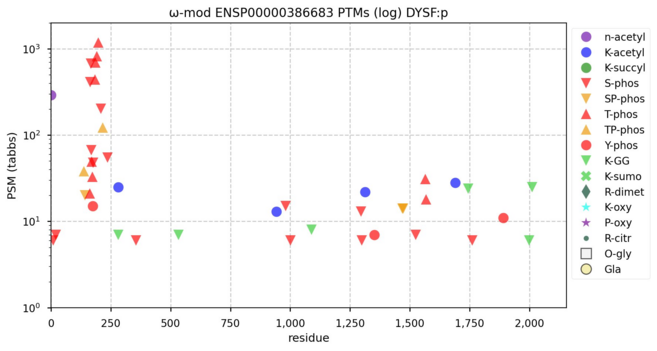
122 followers · 630 posts · Server mastodon.socialModification diagrams for human dysferlin & myoferlin―DYSF:p & MYOF:p―ferlin family subunits where the genes are paralogues but the proteins are not proteologues. #teamProteome
NorSivaeb · @RonBeavis
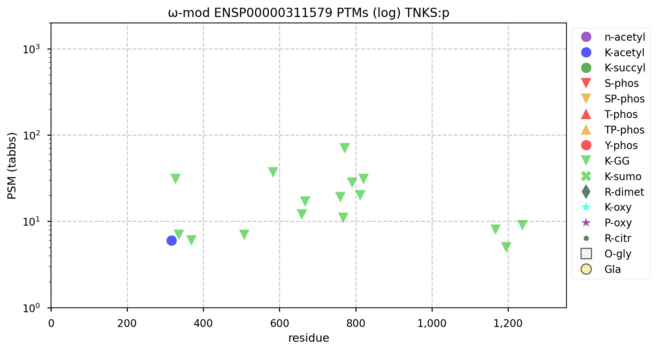
122 followers · 625 posts · Server mastodon.socialModification diagrams for human tankyrase & tankyrase binding protein―TNKS & TNKS1BP1―demonstrating contrasting attitudes towards participation in signalling cascades. #teamProteome
NorSivaeb · @RonBeavis
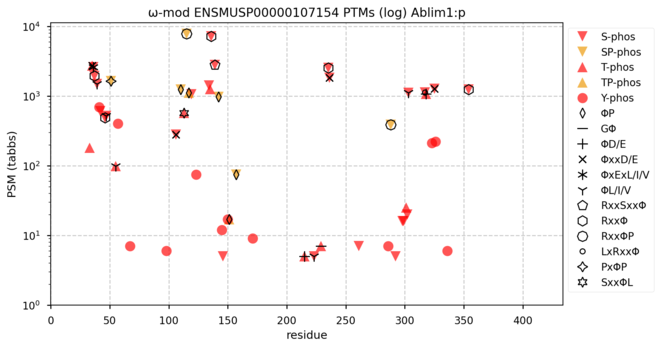
122 followers · 620 posts · Server mastodon.socialS/T/Y phosphorylation diagram for mouse actin-binding LIM protein 1―Ablim1:p―with known gang affiliations. #teamProteome
NorSivaeb · @RonBeavis
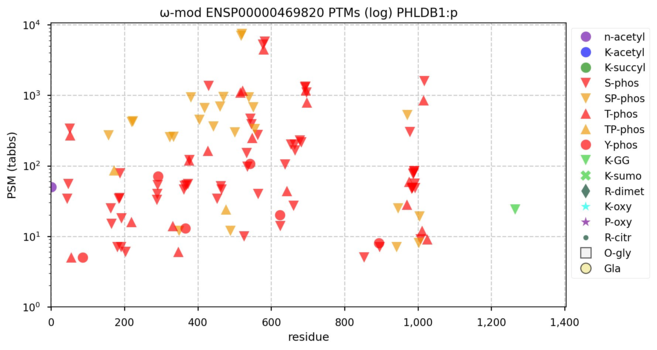
122 followers · 617 posts · Server mastodon.socialω-mod diagrams for human pleckstrin homology like domain family B members 1, 2 & 3―PHLDB1:p, PHLDB2:p, PHLDB3:p―one of these things is not like the others. NB: the sequences of 1 & 2 don't overlap very much, but their PTM patterns really do rhyme. #teamProteome
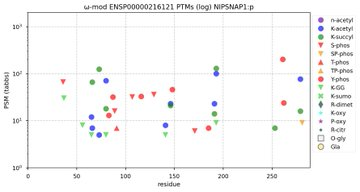
NorSivaeb · @RonBeavis
122 followers · 614 posts · Server mastodon.socialModification diagrams for human NIPSNAP1:p, NIPSNAP2:p, NIPSNAP3A:p & NIPSNAP3B:p. Small, similar mitochondrial matrix proteins of unknown function with rather blah PTMs. #teamProteome
NorSivaeb · @RonBeavis
122 followers · 606 posts · Server mastodon.socialModification & N-linked glycosylation diagrams for human multimerin 1―MMRN1:p―lots of glycosylation, although why is one for the future. #teamProteome
NorSivaeb · @RonBeavis
122 followers · 600 posts · Server mastodon.socialNeed the proteomics version of a "Field Guide to Identifying Cell Lines". #teamProteome