Stephen Matheson🌵🌲 · @sfmatheson
709 followers · 674 posts · Server fediscience.orgNow Golnaz Vahedi from UPenn
https://vahedilab.com/
speaking on
"Modeling Type 1 #Diabetes progression from single-cell #transcriptomic measurements in #human #islets
Vision is to do #SingleCell profiling of #pancreatic cells and peripheral blood, simultaneously, to identify patients who can benefit from early interventions
#cssinglecells23 #pancreatic #SingleCell #islets #human #transcriptomic #diabetes
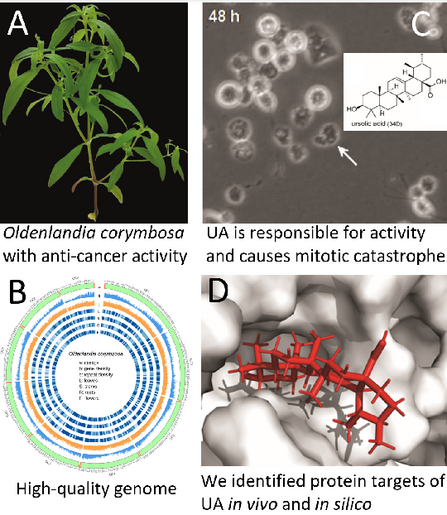
JIPB · @JIPB
42 followers · 180 posts · Server mstdn.science🔓⬇️#OpenAccess #PlantScience for your weekend!
#Genomic, #transcriptomic, and #metabolomic analysis of Oldenlandia corymbosa reveals the #biosynthesis and mode of action of anti-#cancer metabolites.
https://doi.org/10.1111/jipb.13469
@wileyplantsci
#JIPB #PlantSci #medicine #PlantScience
#openaccess #plantscience #genomic #transcriptomic #metabolomic #biosynthesis #cancer #JIPB #plantsci #medicine
CellBioNews · @cellbionews
61 followers · 1480 posts · Server scientificnetwork.de#Cochlea #cell atlas built from single-cell sequencing discovers new cell types, uncovers hidden molecular features.
#genomics #transcriptomic #Emilin2 #mRNA
https://phys.org/news/2023-06-cochlea-cell-atlas-built-single-cell.html
#cochlea #cell #genomics #transcriptomic #emilin2 #mRNA
Christos Argyropoulos MD, PhD · @ChristosArgyrop
317 followers · 412 posts · Server mstdn.scienceAnd this simulation #rstats code, shows why one must use use very short seeds when using #blast to map #nanopore #transcriptomic #RNAseq data (Q10 quality scores are not unheard of, in such datasets).
Take home message, one has got to work for their database searches for long read platforms
(R code in the alt description)
#rstats #blast #nanopore #transcriptomic #rnaseq
CellBioNews · @cellbionews
50 followers · 1164 posts · Server scientificnetwork.deNew method developed to infer #gene #regulatory networks from single-#cell #transcriptomic data.
https://phys.org/news/2023-04-method-infer-gene-regulatory-networks.html
#gene #Regulatory #cell #transcriptomic
Christos Argyropoulos MD, PhD · @ChristosArgyrop
312 followers · 374 posts · Server mstdn.scienceAlright, #bioinformatics folk, which multiple sequence #alignment and #clustering packages should I add to my #transcriptomic @nanopore workflow?
Linclust, uclust/ cd-hit are candidates for clustering.
Candidates for MSA: clustal/kalign/hmmalign.
Other suggestions? #nanopore #sequencing
#bioinformatics #alignment #clustering #transcriptomic #nanopore #sequencing
Gene Ontology · @go
201 followers · 182 posts · Server genomic.socialDon't miss this great #OpenAccess #review in @Non_Coding_RNA about #MicroRNA resources- it has a wonderful section about the contributions and use of the #GeneOntology for examining #transcriptomic data: DOI: 10.3390/ncrna9020018
---
RT @UCLgene
Fantastic to see my previous student, Diana Luna Buitrago, writing this great review of microRNA analysis tools: Insights into Online microRNA Bioinformatics Tools https://www.mdpi.com/2311-553X/9/…
https://twitter.com/UCLgene/status/1632682509069680641
#openaccess #review #MicroRNA #GeneOntology #transcriptomic
TrangAston :verified: · @CastlTrAstonDrs
1192 followers · 2100 posts · Server med-mastodon.com#Cancerbiology #oncologyresearch @naturemedicine Clinical, genomic and #transcriptomic analyses of paired samples of synchronous bilateral female #breastcancer identify associations between tumor concordance and #immune infiltrates levels and response to neoadjuvant treatment @FabienReyal @JoshWaterfall
📌 https://www.nature.com/articles/s41591-023-02216-8
📌 https://www.nature.com/nm/
#cancerbiology #oncologyresearch #transcriptomic #breastcancer #immune
KelvinTuong · @KelvinTuong
42 followers · 80 posts · Server genomic.socialRT @Kelvyinzinho
Delighted to share our latest collab project led by @johwirth with @Meier_Lab. 🤩 #spatial #transcriptomic #DBiTseq
Check it out👇🏼
Spatial transcriptomics using multiplexed deterministic barcoding in tissue | Nature Communications https://www.nature.com/articles/s41467-023-37111-w
#Spatial #transcriptomic #dbitseq
Lucie Bittner · @luciebittner
70 followers · 100 posts · Server ecoevo.socialRT @rmwaterhouse
12 #PhD positions in the LongTREC #European #MarieCurie Network - a programme that will develop methods and tools for the analysis of #transcriptomic data obtained with the most recent single-molecule, long-reads #sequencing technologies.
#phd #european #mariecurie #transcriptomic #sequencing
Rob Waterhouse · @rmwaterhouse
412 followers · 55 posts · Server ecoevo.social12 #PhD positions in the LongTREC #European #MarieCurie Network - a programme that will develop methods and tools for the analysis of #transcriptomic data obtained with the most recent single-molecule, long-reads #sequencing technologies.
#phd #european #mariecurie #transcriptomic #sequencing
Gene Ontology · @go
106 followers · 48 posts · Server genomic.socialAbsolutely fantastic new paper from Marie Brasseur & @leeselab in @BioMedCentral featuring Gammarus fossarum - a wonderful demonstration of the #geneontology in use (thanks for the citation!) @VascoElbrecht @rksalis @weiss_martina @Daniel_G1978 #goannotation
---
RT @leeselab
New study by our @unidue team & @Leibniz_LIB on #multiplestressor impacts lead by Marie Brasseur 🎉. We herein focused on #transcriptomic analysis of the #freshwater k…
https://twitter.com/leeselab/status/1601184358370459648
#GeneOntology #goannotation #multiplestressor #transcriptomic #freshwater
Summer (she/they) · @mellamosummer
23 followers · 1 posts · Server ecoevo.socialMy first #toot & #introduction on Mastodon after soft-exiting #Twitter:
Hey y'all. I'm a 2nd-year #PhD student at #UGA in the #PlantBiology department 🌱. I got my BS in #Botany at #CPP. I'm currently a #NSFGRFP & UGA Presidential fellow.
I research 🌸 #FlowerColor in Geranium & using 🧬 #Transcriptomic, #Phylogenomic, & #CitizenScience approaches.
I'm also really passionate about #Diversity in #STEM as a 🏳️🌈 #Queer & 🇳🇮🇲🇽-🇺🇸 #Latinx student!
I'm hoping to reconnect with my Twitter network here
#toot #introduction #twitter #phd #UGA #PlantBiology #botany #cpp #nsfgrfp #FlowerColor #transcriptomic #phylogenomic #citizenscience #diversity #stem #queer #latinx
Riccardo Baroncelli · @riccardobaroncelli
17 followers · 10 posts · Server genomic.socialGlad to see that our new work: "Comparative #transcriptomic provides novel insights into the #soybean response to #Colletotrichum truncatum infection" is finally out in @FrontPlantSci@twitter.com. Great collaboration between @ESALQMidias@twitter.com, @CIALE_USAL@twitter.com and @Unibo@twitter.com
https://www.frontiersin.org/articles/10.3389/fpls.2022.1046418/full
#transcriptomic #soybean #Colletotrichum
Sarah Salisbury · @SarahSalisbury
121 followers · 74 posts · Server ecoevo.socialEarly view from #MolecularEcology investigating #methylation and #transcriptomic response of #Crassostrea #hongkongensis to #OceanAccidification.
#MolecularEcology #methylation #transcriptomic #crassostrea #hongkongensis #oceanaccidification #aquaculture #Genomics
Alexis Bédécarrats · @alexisbedecarrats
6 followers · 3 posts · Server mastodon.social#introduction I am a young researcher in #neuroscience. I am currently collaborating with Romuald Nargeot at the University of Bordeaux using the mollusk Aplysia as system to study learning and memory. Our aim is to develop and apply single-cell #transcriptomic and epigenetic analysis in a natural organoid whose neurons have been identified and their involvement in behavior and learning established.
#introduction #neuroscience #transcriptomic
Manuel Landesfeind · @landesfeind
94 followers · 149 posts · Server genomic.socialHello #fediverse, here is my #introduction: I am a #Bioinformatician at Evotec and I am here for the #science 😉
From a #work, main interest is in processing, managing and analyzing large #Transcriptomics and #Genomics data sets to boost #DrugDiscovery. My team focuses on data from our #transcriptomic screening platform #ScreenSeq and #multiomics data within the Molecular Patient Database (E.MPD).
Private interest are in #linux, #programming and lately I started to learn and love #Rust 🥰
#Fediverse #introduction #Bioinformatician #science #work #Transcriptomics #genomics #DrugDiscovery #transcriptomic #ScreenSeq #multiomics #linux #programming #rust