PLOS Biology · @PLOSBiology
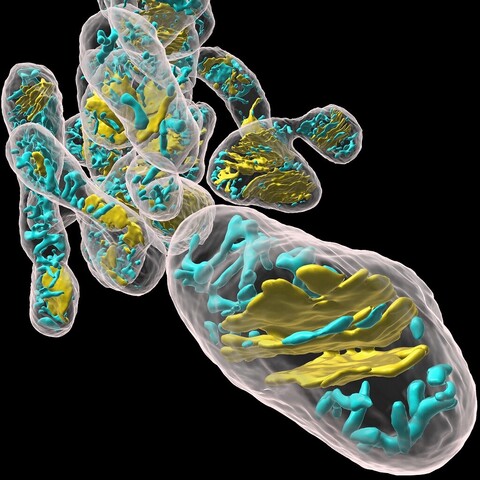
5368 followers · 1706 posts · Server fediscience.orgUnprecedented nanometer-scale structures of cristae in >400 individual #mitochondria, by @Hirabayashi_Lab, using a novel deep learning-based analysis of #volumeEM images, (also reveals previously unknown function of OPA1) #PLOSBiology https://plos.io/3OXtPzK
#plosbiology #volumeem #mitochondria
Albert Cardona · @albertcardona
2023 followers · 3910 posts · Server mathstodon.xyzFollowing from Collinson's paper, here is my take on scaling up volume electron microscopy for connectomics in the #UK or any country willing to commit about 10 to 20 million a year:
"Growing and nurturing a research base in connectomics"
https://albert.rierol.net/tell/20230823_connectomics_research_base.html
From the humble #Drosophila to the #mouse brain, passing through the mosquito, honeybee, gecko lizards and the Etruscan shrew.
It's possible, it's feasible, it's timely: therefore we must.
#vem #volumeem #connectomics #neuroscience #mouse #drosophila #uk
Albert Cardona · @albertcardona
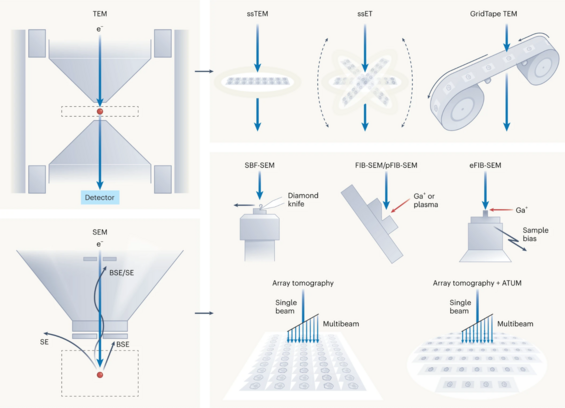
2023 followers · 3906 posts · Server mathstodon.xyz"Volume EM: a quiet revolution takes shape" – a review and commentary by Lucy Collinson et al. 2023 on present and future electron microscopy technology and its application https://www.nature.com/articles/s41592-023-01861-8
#electronmicroscopy #volumeem #vem
Albert Cardona · @albertcardona
2018 followers · 3871 posts · Server mathstodon.xyz
Amusing that the piece includes optical microscopy methods unrelated to neural function, such as STED, PALM, expansion microscopy and CLARITY, but ignores electron microscopy methods key to the current boom in #connectomics. In particular, Denk and Horstmann 2004 for SBEM https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0020329 , Knott et al. 2008 for first application of FIBSEM to neuronal tissue https://www.jneurosci.org/content/28/12/2959 , Schalek et al. 2011 for the ATUM serial section tape-collecting system https://academic.oup.com/mam/article-abstract/17/S2/966/6947317 , Xu et al. 2017 for e-FIBSEM and Graham et al. 2019 for GridTape TEM https://www.biorxiv.org/content/10.1101/657346.abstract
#vem #volumeem #microscopy #neuroscience #connectomics
Stephen Royle · @steveroyle
933 followers · 369 posts · Server biologists.socialFor any #3DEM #volumeEM people: is there an alternative software for skinning that you can recommend?
I’m building a 3D model of a complex subcellular structure, using 3DMOD. During skinning, imodmesh is generating a many misconnections. I've minimised them but I’m wondering if other software has better algorithms for generating the mesh.
FOSS-preferred. Please don't suggest Amira!
Journal of Cell Science · @J_Cell_Sci
603 followers · 50 posts · Server biologists.socialKarel Mocaer, Yannick Schwab, Paolo Ronchi and colleagues reveal the 3D ultrastructure of a marine dinoflagellate using their CLEM workflow for targeting environmental microorganisms from heterogeneous samples.
Highlight: https://journals.biologists.com/jcs/article/136/15/e136_e1502/325835/vCLEM-reveals-the-ultrastructure-of-phytoplankton
T&R: https://journals.biologists.com/jcs/article/136/15/jcs261355/325830/Targeted-volume-correlative-light-and-electron
Find out more about this research
in our ‘First person’ interview with Karel Mocaer: https://journals.biologists.com/jcs/article/136/15/jcs261499/325833/First-person-Karel-Mocaer
#openaccess #ReadandPublish #cellbiology #volumeEM @ReviewCommons
#openaccess #readandpublish #cellbiology #volumeem
Josh Moore · @joshmoore
503 followers · 252 posts · Server fediscience.orgAnd it begins... the 1st #GordonConf on #volumeEM!
Thanks for the invitation to #vEMtura23 🤩🌅🔬🧊
#vemtura23 #volumeem #gordonconf
FocalPlane · @focalplane_jcs
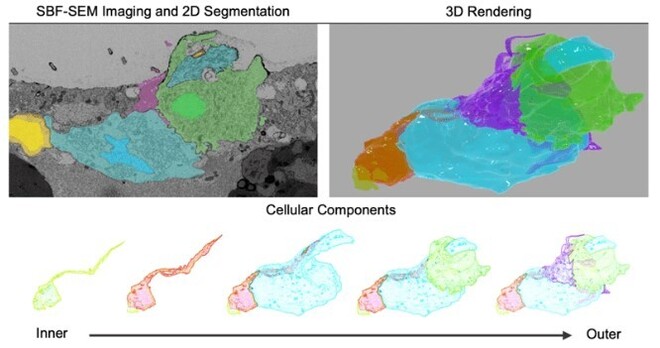
405 followers · 18 posts · Server biologists.socialThis #EMmonday we’re highlighting the 3D architecture of the zebrafish germline cyst by SBF-SEM from Vineet Kumar & Yaniv Elkouby. Check out their materials and methods for the manual segmentation protocol using TrakEM2.
https://journals.biologists.com/dev/article/150/13/dev201349/321182/Tools-to-analyze-the-organization-and-formation-of
#emmonday #microscopy #cellbiology #volumeem
Royle Lab News · @roylelab
129 followers · 47 posts · Server mstdn.scienceNew lab paper! Bio-protocol from @NFerrandiz
3D Ultrastructural Visualization of Mitosis Fidelity in Human Cells Using Serial Block Face Scanning Electron Microscopy (SBF-SEM)
#cellbiology #imaging #volumeem
Albert Cardona · @albertcardona
1860 followers · 3156 posts · Server mathstodon.xyzBack in 2015 when we published our first work featuring a connectome, many in the #Drosophila community told me, now you’ve raised the bar, further functional and behavioural studies on the nervous system will have to be on the basis of #connectomics.
And indeed the contrast was immediately stark between studies based on the connectome and studies in the old style of a “circuit” consisting of a stimulus, a GAL4 line, and a behavioural readout; the latter appeared obsolete overnight. A bit afterwards Larry Abbott coined the new BC/AC divide: before and after the connectome. Because the connectome isn’t sufficient but it is necessary for progress in understanding brain function.
Drosophila #neuroscience is well into the AC era. #Zebrafish is very close to entering—#VolumeEM for the whole larval zebrafish already exists and dimensions are tractable with current technology. One wonders whether the effect will spread onto mouse studies soon; there, the cost of mapping the connectome is presently measured in billions of dollars. Amusingly, the mouse field is hiring heavily from fly labs. I’m cheering for them all.
#volumeem #zebrafish #neuroscience #connectomics #drosophila
Albert Cardona · @albertcardona
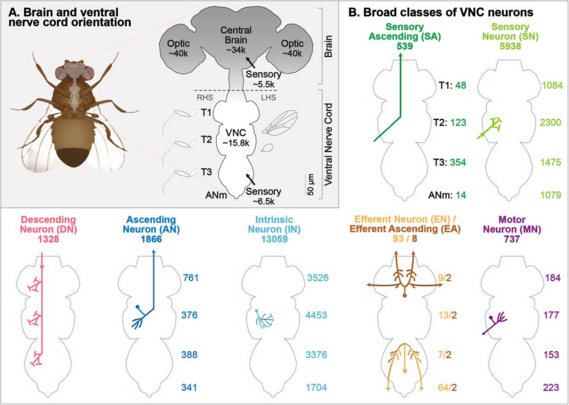
1846 followers · 3077 posts · Server mathstodon.xyz"Systematic annotation of a complete adult male Drosophila nerve cord connectome reveals principles of functional organisation", by Marin et al. 2023 (Greg Jefferis lab at #MRCLMB in collaboration with #HHMIJanelia and others).
https://www.biorxiv.org/content/10.1101/2023.06.05.543407v1
On the analysis of the #Drosophila male adult nerve cord (#MANC), reconstructed from #VolumeEM by Takemura et al. 2023 (preprint still pending to come out).
#connectomics #neuroscience #volumeem #manc #drosophila #HHMIJanelia #MRCLMB
FocalPlane · @focalplane_jcs
362 followers · 183 posts · Server mstdn.scienceThis #EMmonday we are focussing on SBF-SEM with a case study from Federica Mangione. Federica and colleagues used SBF-SEM to dissect the cellular assembly of tactile organ of Drosophila, identifying a new cell type, the F-Cell.
https://focalplane.biologists.com/2023/05/19/inputs-and-outputs-of-vem-in-a-sensory-system/
For more from the volume EM community: https://focalplane.biologists.com/category/volume-em/
#emmonday #volumeem #drosophila #DevBio
Albert Cardona · @albertcardona
1805 followers · 2680 posts · Server mathstodon.xyz@BorisBarbour @alexh @JeffZemla
Question is who pays for it, and for how long.
The data is acquired at 16-bit, yet often served at 8-bit (2x reduction), and further compressed with either lossless (~20% further savings) or lossy (~66% further savings) formats to speed up on-demand data transfer. I'm talking about #VolumeEM data. So a ~20TB data set becomes under 4TB: still way too large for publishers to be willing to host in perpetuity alongside the paper.
Relatedly, I use the Rubin razor to decide whether a data set is worth keeping around: is the cost of storage lower than the cost of reacquiring/remaking the data set, given new methods? So far it's always been cheaper to store it, but within our own servers.
FocalPlane · @focalplane_jcs
359 followers · 153 posts · Server mstdn.science📢 New post on FocalPlane
Check out the #volumeEM case study from Federica Mangione on serial blockface SEM for dissecting the cellular assembly of tactile bristles in Drosophila.
https://focalplane.biologists.com/2023/05/19/inputs-and-outputs-of-vem-in-a-sensory-system/
Kurianlab · @kurianlab
83 followers · 637 posts · Server sciencemastodon.comRT @manorlaboratory
Honored to have played a part in this insane tour de force effort from Nazma Malik in the Reuben Lab @LabReuben. You’ll have to dig around, but if you do you’ll find some gorgeous 😍🤯 #VolumeEM highlighting mitochondria-lysosome contacts https://www.science.org/doi/10.1126/science.abj5559
BioImagingUK · @BioImagingUK
41 followers · 222 posts · Server mstdn.scienceKeep your 👀 on #VolumeEM, it's a Nature method to watch in 2023 and this paper introduces the growing @VolumeEM1 community. New members are very welcome!!
---
RT @naturemethods
Out today from Collinson, Verkade, and colleagues, an exciting Comment describing the "quiet revolution" that has been taking shape in volume EM and the grassroots community effort to build and disseminate these technologies @EM_STP @PaulVerkade @VolumeEM1 htt…
https://twitter.com/naturemethods/status/1648719004687650816
BioImagingUK · @BioImagingUK
41 followers · 221 posts · Server mstdn.scienceIt's out! @naturemethods paper all about the @VolumeEM1 community initiative -
aka "New Kid on the Blockface!" http://nature.com/articles/s4159…
#VolumeEM
@EM_STP @PaulVerkade @JemimaBurden @Trickymons @AnwenBullen @MDarrow7 @KedarNarayan2 @caraffa62 @martinjones78 @connectomx3d
---
RT @VolumeEM1
*🔥🔥🔥Hot off the press🔥🔥🔥*
Volume EM: a quiet revolution takes shape
#VolumeEM
https://www.nature.com/articles/s41592-023-01861-8
https://twitter.com/VolumeEM1/status/1648714331549253634
BioImagingUK · @BioImagingUK
41 followers · 218 posts · Server mstdn.scienceIt's out! @naturemethods paper all about the @VolumeEM1 community initiative -
aka "New Kid on the Blockface!" https://www.nature.com/articles/s41592-023-01861-8
#VolumeEM
@EM_STP @PaulVerkade @JemimaBurden @Trickymons @AnwenBullen @MDarrow7 @KedarNarayan2 @caraffa62 @martinjones78 @connectomx3d
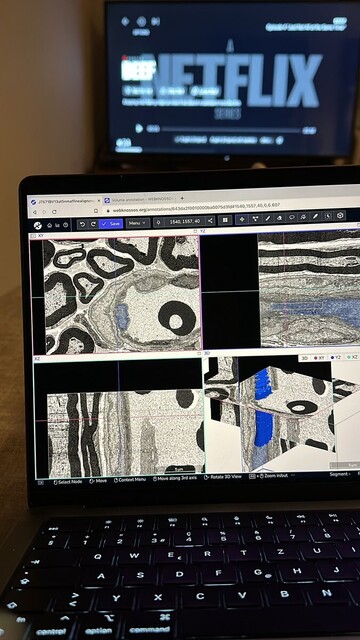
WEBKNOSSOS · @webknossos
84 followers · 74 posts · Server mstdn.scienceBioImagingUK · @BioImagingUK
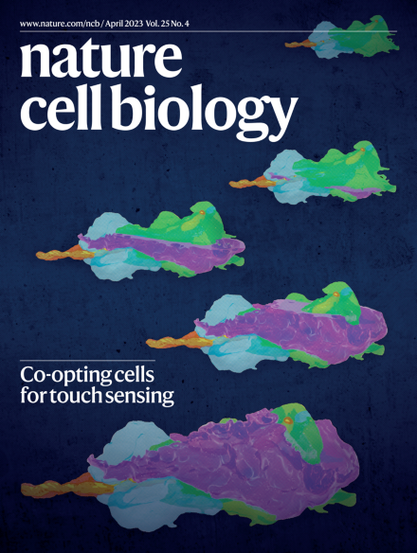
41 followers · 211 posts · Server mstdn.scienceGreat to see #volumeEM on the cover of Nature Cell Biology! Follow @VolumeEM1 to keep up to date with the community!
---
RT @taponlab
If you missed the story of the F-cell by Mangione et al., catch it on the cover of @NatureCellBio and read the highlight, https://www.nature.com/articles/s41556-023-01119-7. We are grateful for the thoughtful highlight-and congrats to Fede and Catherine (@EM_STP) for the cover image!
https://twitter.com/taponlab/status/1646976774415302657